Abstract
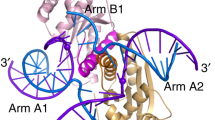
A single enzyme active site that catalyzes multiple reactions is a well-established biochemical theme, but how one nuclease site cleaves both DNA strands of a double helix has not been well understood. In analyzing site-specific DNA cleavage by the mammalian RAG1–RAG2 recombinase, which initiates V(D)J recombination, we find that the active site is reconfigured for the two consecutive reactions and the DNA double helix adopts drastically different structures. For initial nicking of the DNA, a locally unwound and unpaired DNA duplex forms a zipper via alternating interstrand base stacking, rather than melting as generally thought. The second strand cleavage and formation of a hairpin–DNA product requires a global scissor-like movement of protein and DNA, delivering the scissile phosphate into the rearranged active site.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The accession numbers for the cryo-EM structures and associated density maps of the mouse PRC and NFC complexes reported in this paper have been deposited to the PDB and EMDB under accession codes PDB 6OEM to 6OER and 6V0V and EMD-20030 to EMD-20035, EMD-20038, EMD-20039 and EMD-21003, as specified in Table 1.
References
Mizuuchi, K. Transpositional recombination: mechanistic insights from studies of Mu and other elements. Annu. Rev. Biochem. 61, 1011–1051 (1992).
Gellert, M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 71, 101–132 (2002).
Schatz, D. G. & Swanson, P. C. V(D)J recombination: mechanisms of initiation. Annu. Rev. Genet. 45, 167–202 (2011).
Kim, M. S., Lapkouski, M., Yang, W. & Gellert, M. Crystal structure of the V(D)J recombinase RAG1–RAG2. Nature 518, 507–511 (2015).
Sakano, H., Huppi, K., Heinrich, G. & Tonegawa, S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature 280, 288–294 (1979).
Lewis, S. M. The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv. Immunol. 56, 27–150 (1994).
Lapkouski, M., Chuenchor, W., Kim, M. S., Gellert, M. & Yang, W. Assembly pathway and characterization of the RAG1/2–DNA paired and signal-end complexes. J. Biol. Chem. 290, 14618–14625 (2015).
Ru, H. et al. Molecular mechanism of V(D)J recombination from synaptic RAG1–RAG2 complex structures. Cell 163, 1138–1152 (2015).
Kim, M. S. et al. Cracking the DNA code for V(D)J recombination. Mol. Cell 70, 358–370 (2018).
Ru, H. et al. DNA melting initiates the RAG catalytic pathway. Nat. Struct. Mol. Biol. 25, 732–742 (2018).
Davies, D. R., Goryshin, I. Y., Reznikoff, W. S. & Rayment, I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science 289, 77–85 (2000).
Richardson, J. M., Colloms, S. D., Finnegan, D. J. & Walkinshaw, M. D. Molecular architecture of the Mos1 paired-end complex: the structural basis of DNA transposition in a eukaryote. Cell 138, 1096–1108 (2009).
Hare, S., Gupta, S. S., Valkov, E., Engelman, A. & Cherepanov, P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464, 232–236 (2010).
Montano, S. P., Pigli, Y. Z. & Rice, P. A. The Mu transpososome structure sheds light on DDE recombinase evolution. Nature 491, 413–417 (2012).
Hickman, A. B. et al. Structural basis of hAT transposon end recognition by Hermes, an octameric DNA transposase from Musca domestica. Cell 158, 353–367 (2014).
Passos, D. O. et al. Cryo-EM structures and atomic model of the HIV-1 strand transfer complex intasome. Science 355, 89–92 (2017).
Yusa, K. piggyBac transposon. Microbiol. Spectr. 3, MDNA3-0028-2014 (2015).
Lesbats, P., Engelman, A. N. & Cherepanov, P. Retroviral DNA integration. Chem. Rev. 116, 12730–12757 (2016).
Nowotny, M., Gaidamakov, S. A., Crouch, R. J. & Yang, W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell 121, 1005–1016 (2005).
Grundy, G. J., Yang, W. & Gellert, M. Autoinhibition of DNA cleavage mediated by RAG1 and RAG2 is overcome by an epigenetic signal in V(D)J recombination. Proc. Natl Acad. Sci. USA 107, 22487–22492 (2010).
Grundy, G. J., Hesse, J. E. & Gellert, M. Requirements for DNA hairpin formation by RAG1/2. Proc. Natl Acad. Sci. USA 104, 3078–3083 (2007).
Mills, J. B. & Hagerman, P. J. Origin of the intrinsic rigidity of DNA. Nucleic Acids Res. 32, 4055–4059 (2004).
Yakovchuk, P., Protozanova, E. & Frank-Kamenetskii, M. D. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 34, 564–574 (2006).
Zgarbova, M. et al. Refinement of the Cornell et al. nucleic acids force field based on reference quantum chemical calculations of glycosidic torsion profiles. J. Chem. Theory Comput. 7, 2886–2902 (2011).
Hart, K. et al. Optimization of the CHARMM additive force field for DNA: improved treatment of the BI/BII conformational equilibrium. J. Chem. Theory Comput. 8, 348–362 (2012).
Rubio-Cosials, A. et al. Transposase–DNA complex structures reveal mechanisms for conjugative transposition of antibiotic resistance. Cell 173, 208–220 (2018).
Atkinson, P. W. hAT transposable elements. Microbiol. Spectr. 3, MDNA3-0054-2014 (2015).
Ru, H., Zhang, P. & Wu, H. Structural gymnastics of RAG-mediated DNA cleavage in V(D)J recombination. Curr. Opin. Struct. Biol. 53, 178–186 (2018).
Hickman, A. B. et al. Structural insights into the mechanism of double strand break formation by Hermes, a hAT family eukaryotic DNA transposase. Nucleic Acids Res. 46, 10286–10301 (2018).
Ramsden, D. A., McBlane, J. F., van Gent, D. C. & Gellert, M. Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J. 15, 3197–3206 (1996).
Hesse, J. E., Lieber, M. R., Mizuuchi, K. & Gellert, M. V(D)J recombination: a functional definition of the joining signals. Genes Dev. 3, 1053–1061 (1989).
Hu, J. et al. Chromosomal loop domains direct the recombination of antigen receptor genes. Cell 163, 947–959 (2015).
Nowotny, M. Retroviral integrase superfamily: the structural perspective. EMBO Rep. 10, 144–151 (2009).
Yuan, Y. W. & Wessler, S. R. The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc. Natl Acad. Sci. USA 108, 7884–7889 (2011).
Boboila, C., Alt, F. W. & Schwer, B. in Adv. Immunol Vol 116 (ed. Alt, F. A.) Ch. 1 (Elsevier, 2012).
Deriano, L. & Roth, D. B. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu. Rev. Genet. 47, 433–455 (2013).
Grundy, G. J. et al. Initial stages of V(D)J recombination: the organization of RAG1/2 and RSS DNA in the postcleavage complex. Mol. Cell 35, 217–227 (2009).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Fernandez-Leiro, R. & Scheres, S. H. W. A pipeline approach to single-particle processing in RELION. Acta Crystallogr. D Struct. Biol. 73, 496–502 (2017).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Bai, X. C., Rajendra, E., Yang, G., Shi, Y. & Scheres, S. H. Sampling the conformational space of the catalytic subunit of human γ-secretase. Elife 4, e11182 (2015).
Swint-Kruse, L. & Brown, C. S. Resmap: automated representation of macromolecular interfaces as two-dimensional networks. Bioinformatics 21, 3327–3328 (2005).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Barad, B. A. et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015).
Páll, S., Abraham, M. J., Kutzner, C., Hess, B. & Lindahl, E. in Solving Software Challenges for Exascale (eds Markidis, S. & Laure, E.) 3–27 (Lecture Notes in Computer Science 8759, Springer, 2015).
Tribello, G. A., Bonomi, M., Branduardi, D., Camilloni, C. & Bussi, G. PLUMED2: new feathers for an old bird. Comput. Phys. Commun. 185, 604–613 (2014).
Bussi, G., Donadio, D. & Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 (2007).
Parrinello, M. & Rahman, A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981).
Domanski, J., Sansom, M. S. P., Stansfeld, P. J. & Best, R. B. Balancing force field protein–lipid interactions to capture transmembrane helix–helix association. J. Chem. Theory Comput. 14, 1706–1715 (2018).
Grosse-Kunstleve, R. W. & Adams, P. D. Substructure search procedures for macromolecular structures. Acta Crystallogr. D Biol. Crystallogr. 59, 1966–1973 (2003).
McCoy, A. J., Storoni, L. C. & Read, R. J. Simple algorithm for a maximum-likelihood SAD function. Acta Crystallogr. D Biol. Crystallogr. 60, 1220–1228 (2004).
Acknowledgements
This research was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (M.G., DK036167; W.Y., DK036147 and DK036144; Z.H.Z., GM071940). The authors acknowledge the use of instruments at the Electron Imaging Center for NanoMachines supported by the NIH (1S10RR23057, 1S10OD018111 and U24GM116792), NSF (DBI-1338135 and DMR-1548924) and CNSI at UCLA.
Author information
Authors and Affiliations
Contributions
X.C. carried out all experiments and structure determination. Y.C. collected cryo-EM micrographs on the Krios microscope at UCLA and helped with structure determination and refinement. H.W. helped with cryo-EM data collection on the TF20 and Krios at NIH. R.B.B. carried out molecular dynamics simulations. Z.H.Z., W.Y. and M.G. supervised the research project. X.C., R.B.B., W.Y. and M.G. prepared the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Anke Sparmann was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Cleavage and cryo-EM analysis of DNA substrates in NFC.
a, b, Cleavage efficiencies (nicking and hairpinning) of the three DNA variants by WT mRAG at 22 and 37 °C (mean and s.d., n= 3 independent samples). c, Percentage of NFC and PRC (NFC/PRC) in cryo-EM 3D classification from samples made of DNA0, DNA1 or DNA2 substrate with E962Q mutant mRAG at 22 and 37 °C. Asterisk (*) indicates that the dataset was collected on a Tecnai F20 electron microscope instead of Titan Krios.
Extended Data Fig. 2 Structure determination of mouse NFC with DNA1 by cryo-EM.
a, Flow chart for cryo-EM data processing of mRAG complexed with DNA1. The maps with red bold letter are used for final model building. b, A surface presentation of the 3.7 Å NFC (DNA1) map (C1 symmetry). Colors are according to the local resolution estimated by ResMap, and the color scale bar is shown on its right. c, Angular distributions of all particles used for the final three-dimensional reconstruction shown in b. d, The FSC curves of the NFC (DNA1) map (C1). The “gold standard” FSC between two independent halves of the map (black line) indicates a resolution of 3.7 Å, and the blue line is the FSC between the final refined model and the final map. e to i, Representative regions of the C1 map (transparent grey surface). The maps are shown with the final structural models (cartoon or stick) superimposed.
Extended Data Fig. 3 Structure determination of mouse NFC with DNA2 by cryo-EM.
a, Flow chart for cryo-EM data processing of mRAG complexed with DNA2. The maps with red bold letter are used for final model building. b, A surface presentation of the 3.3 Å NFC (DNA2) map (C1 symmetry). Colors are according to the local resolution estimated by ResMap, and the color scale bar is shown on its right. c, Angular distributions of all particles used for the final three-dimensional reconstruction shown in b. d, The FSC curves of NFC (DNA2) map (C1). The “gold standard” FSC between two independent halves of the map (black line) indicates a resolution of 3.3 Å, and the blue line is the FSC between the final refined model and the final map. e to i, Representative regions of the C1 map (transparent grey surface). The maps are shown with the final structural models (cartoon or stick) superimposed.
Extended Data Fig. 4 Structural comparisons of mouse PRC and NFC.
a, Cryo-EM structures of NFC with DNA1 (green) and DNA2 (blue) are superimposable. b, Comparison of mouse PRC and NFC structures. Superposition of cryo-EM PRC (red) and NFC (DNA1) (green) structures reveals limited NBD and nonamer movement, which is marked with blue dashed circle (right panel). c, Superposition of crystal (grey) and cryo-EM (red) PRC structures reveals the different NBD and nonamer region (circled in red dashes) due to crystal-lattice contacts. d, e, The zoom-in views of the active center and DNA distortions in the superimposed structures shown in a-b. The catalytic DDE motif and two metal ions (a and b) are labeled; the heptamer of RSS DNA is shown in detailed cartoon presentation; the scissile phosphate of top strand is marked by a large ball. In panel d, bases forming the DNA zipper are labeled. f, A zoom-in view of boxed area in panel b. RAG2 interacts with the minor groove in PRC or the major groove in NFC.
Extended Data Fig. 5 Structure determination of mouse WT NFC with DNA0 by cryo-EM.
a, Flow chart for cryo-EM data processing of WT mRAG complexed with DNA0. Data processing was done using RELION. The 3.6Å map labeled in red was used for final model building. b, A surface presentation of the 3.6 Å map of WT NFC (DNA0). Colors are according to the local resolution estimated by ResMap, and the color scale bar is shown on its right. c, Angular distributions of all particles used for the final three-dimensional reconstruction. d, The FSC curves of WT NFC (DNA0) map. The “gold standard” FSC between two independent halves of the map indicates an overall resolution of 3.6 Å. e–h, Representative regions of the map (transparent grey surface). The refined zipper DNA fits the map better (e) than the melted DNA (PDB: 6DBR) (f).
Extended Data Fig. 6 Molecular simulations of the untwisted region of DNA.
a, b, Unbiased simulations were run starting from the base-flipped out structure (PDB 6DBR) (a), and the zipper structure (b) using the Amber 14 force field. Plotted in each case are the all-atom RMSD to the base-flipped out structure (black) and the zippered structure (red). The structures at the start and end of each run are shown above the RMSD plots. c, d, The analogous results are given for simulations with the CHARMM 36 force field. e, Biased simulations were run from a canonical B-DNA form, in which the terminal residues were driven to mimic the stretched and untwisted DNA observed in the mouse and zebrafish NFC structures. The RMSD and initial and final structures are shown as before.
Extended Data Fig. 7 Structure determination of mouse PRC with DNA0 by cryo-EM.
a, Flow chart for the cryo-EM data processing of mRAG complexed with DNA0. The maps with red bold letter are used for final model building. b, A surface presentation of the 3.6 Å NFC map (C1 symmetry). Colors are according to the local resolution estimated by ResMap, and the color scale bar is shown on its right. c, Angular distributions of all particles used for the final three-dimensional reconstruction. d, The FSC curves of PRC (DNA0) map (C1). The “gold standard” FSC between two independent halves of the map (black line) indicates a resolution of 3.6 Å, and the blue line is the FSC between the final refined model and the final map. e to i, Representative regions of the C1 map (transparent grey surface). The maps are shown with the final structural models (cartoon or stick) superimposed. j, The maps of 23RSS in PRC (DNA0), NFC (DNA1) and NFC (DNA2).
Extended Data Fig. 8 Remodeling of the active site in NFC and HFC.
a, Repositioning of αX in the RNH domain during PRC to NFC transition (both are cryo-EM structures). E962 is far from the active site in PRC (light blue) but is positioned for catalysis in NFC (green). b, Anomalous X-ray scattering of the PRC crystals confirms that one Mn2+ and one Zn2+ are bound to each RAG1 subunit. The anomalous map is contoured at 3σ in red. The blue 2Fo-Fc map (contoured at 1σ) highlights R848, which is buried in the minor groove. c, The reconfigured E962 in HFC (pink) after the first DNA cleavage by nicking (green).
Supplementary information
Supplementary Information
Supplementary Fig. 1 and Supplementary Table 1.
Supplementary Video 1
DNA zipper formation. The two base pairs (AC/(TG)) undergoing zipper formation are shown as sticks, and the rest of the DNA is simplified as a tube and ladder cartoon. The active site DDE are shown in red sticks, and divalent cations are represented as green spheres. The second Mg2+ ion appears only at the end because it binds in a fully formed active site with the substrate in place, but not in PRC. The red sphere on the DNA strand marks the scissile phosphate for nicking, and the DNA becomes nicked when two Mg2+ ions are properly bound.
Supplementary Video 2
Morphing of structural transition from PRC to NFC. The mRAG protein is shown as a semitransparent molecular surface. ZnH2 domains (green and light blue cartoon) of RAG1 move significantly and two L12 loops (green and light blue coils) become extended. The second metal ion (green sphere) is captured in the active site (marked by D600, D708 and E962 in red sticks) only when E962 and the scissile phosphate are in the active configuration. The 12/23RSS DNAs are shown as yellow and orange tube-and-ladders.
Supplementary Video 3
Morphing of the structural transition from NFC to HFC. One RAG1–RAG2 heterodimer (bound to 12RSS, yellow) is superimposed between the two structures and shown as a semitransparent molecular surface. The second RAG1–RAG2 heterodimer (bound to 23RSS, orange), shown as a colored cartoon, moves towards RAG-12RSS, delivering the bottom strand on each RSS DNA into the active site. Two L12 loops (green and light blue coil) of RAG1 subunits move away from the RAG1 interface. The configuration of the active center (red stick-and-balls for catalytic residues), except for E962, remains unchanged, and the closing motion positions the scissile phosphates on the bottom strands into the active centers. The green spheres represent Mg2+ ions. The second Mg2+ ion appears only when the active site is fully formed, but not when DNA undergoes conformational changes. When both Mg2+ ions are properly bound in the active site, the DNA cleavage/transesterification reaction takes place as shown.
Supplementary Video 4
The reaction cycle of DNA nicking and hairpin formation catalyzed by RAG recombinase. The scissile phosphates for nicking and hairpinning of DNA1 are highlighted by red and pink spheres. The active site DDE are shown as red sticks, and two metal ions in the NFC and HFC states are shown as green spheres. When the catalytic carboxylates DDE are not fully aligned or the scissile phosphate is not captured in the active site, such as during conformational changes of RAG and DNA, only one metal ion may be retained.
Rights and permissions
About this article
Cite this article
Chen, X., Cui, Y., Best, R.B. et al. Cutting antiparallel DNA strands in a single active site. Nat Struct Mol Biol 27, 119–126 (2020). https://doi.org/10.1038/s41594-019-0363-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-019-0363-2
This article is cited by
-
Structural and biochemical basis for DNA and RNA catalysis by human Topoisomerase 3β
Nature Communications (2022)
-
Structural insights into the evolution of the RAG recombinase
Nature Reviews Immunology (2022)
-
Dimers of DNA-PK create a stage for DNA double-strand break repair
Nature Structural & Molecular Biology (2021)
-
Clinical Manifestations, Mutational Analysis, and Immunological Phenotype in Patients with RAG1/2 Mutations: First Cases Series from Mexico and Description of Two Novel Mutations
Journal of Clinical Immunology (2021)
-
The molecular basis and disease relevance of non-homologous DNA end joining
Nature Reviews Molecular Cell Biology (2020)