Abstract
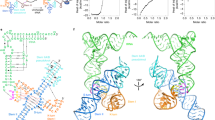
T-box riboregulators are a class of cis-regulatory RNAs that govern the bacterial response to amino acid starvation by binding, decoding and reading the aminoacylation status of specific transfer RNAs. Here we provide a high-resolution crystal structure of a full-length T-box from Mycobacterium tuberculosis that explains tRNA decoding and aminoacylation sensing by this riboregulator. Overall, the T-box consists of decoding and aminoacylation sensing modules bridged by a rigid pseudoknot structure formed by the mid-region domains. Stem-I and the Stem-II S-turn assemble a claw-like decoding module, while the antiterminator, Stem-III, and the adjacent linker form a tightly interwoven aminoacylation sensing module. The uncharged tRNA is selectively recognized by an unexpected set of favorable contacts from the linker region in the aminoacylation sensing module. A complex structure with a charged tRNA mimic shows that the extra moiety dislodges the linker, which is indicative of the possible chain of events that lead to alternative base-pairing and altered expression output.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Atomic coordinates and structure factors for Mtb-ileS_tRNA-OH_native and Mtb-ileS_tRNA-cP_native have been deposited in the Protein Data Bank with the accession codes 6UFG and 6UFH, respectively. Source data for Figs. 2b and 3f are available in Supplementary Table 3. Other data are available upon reasonable request.
References
Mandal, M. & Breaker, R. R. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 5, 451–463 (2004).
Peselis, A. & Serganov, A. Themes and variations in riboswitch structure and function. Biochim. Biophys. Acta 1839, 908–918 (2014).
Grundy, F. J. & Henkin, T. M. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell 74, 475–482 (1993).
Grundy, F. J., Winkler, W. C. & Henkin, T. M. tRNA-mediated transcription antitermination in vitro: Codon-anticodon pairing independent of the ribosome. Proc. Natl Acad. Sci. USA 99, 11121–11126 (2002).
Grigg, J. C. et al. T box RNA decodes both the information content and geometry of tRNA to affect gene expression. Proc. Natl Acad. Sci. USA 110, 7240–7245 (2013).
Zhang, J. & Ferré-D’Amaré, A. R. Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature 500, 363–366 (2013).
Grigg, J. C. & Ke, A. Structural determinants for geometry and information decoding of tRNA by T Box Leader RNA. Structure 21, 2025–2032 (2013).
Grundy, F. J., Rollins, S. M. & Henkin, T. M. Interaction between the acceptor end of tRNA and the T box stimulates antitermination in the Bacillus subtilis tyrS gene: a new role for the discriminator base. J. Bacteriol. 176, 4518–4526 (1994).
Zhang, J. & Ferré-D’Amaré, A. R. Direct evaluation of tRNA aminoacylation status by the T-box riboswitch using tRNA-mRNA stacking and steric readout. Mol. Cell 55, 148–155 (2014).
Grundy, F. J. & Henkin, T. M. Kinetic analysis of tRNA-directed transcription antitermination of the Bacillus subtilis glyQS gene in vitro. J. Bacteriol. 186, 5392–5399 (2004).
Suddala, K. C. et al. Hierarchical mechanism of amino acid sensing by the T-box riboswitch. Nat. Commun. 9,1896 (2018).
Zhang, J. et al. Specific structural elements of the T-box riboswitch drive the two-step binding of the tRNA ligand. eLife 7, e39518 (2018).
Seliverstov, A. V., Putzer, H., Gelfand, M. S. & Lyubetsky, V. A. Comparative analysis of RNA regulatory elements of amino acid metabolism genes in Actinobacteria. BMC Microbiol. 5, 54 (2005).
Sherwood, A. V., Grundy, F. J. & Henkin, T. M. T box riboswitches in Actinobacteria: translational regulation via novel tRNA interactions. Proc. Natl Acad. Sci. USA 112, 1113–1118 (2015).
Rollins, S. M., Grundy, F. J. & Henkin, T. M. Analysis of cis-acting sequence and structural elements required for antitermination of the Bacillus subtilis tyrS gene. Mol. Microbiol. 25, 411–421 (1997).
Saad, N. Y. et al. Two-codon T-box riboswitch binding two tRNAs. Proc. Natl Acad. Sci. USA 110, 12756–12761 (2013).
Shepherd, J. & Ibba, M. Bacterial transfer RNAs. FEMS Microbiol. Rev 39, 280–300 (2015).
Agris, P. F., Vendeix, F. A. P. & Graham, W. D. tRNA’s wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 366, 1–13 (2007).
Putzer, H., Condon, C., Brechemier-Baey, D., Brito, R. & Grunberg-Manago, M. Transfer RNA-mediated antitermination in vitro. Nucleic Acids Res. 30, 3026–3033 (2002).
Ogle, J. M. et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902 (2001).
Ogle, J. M., Murphy, F. V., Tarry, M. J. & Ramakrishnan, V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111, 721–732 (2002).
Demeshkina, N., Jenner, L., Westhof, E., Yusupov, M. & Yusupova, G. A new understanding of the decoding principle on the ribosome. Nature 484, 256–259 (2012).
Vitreschak, A. G., Mironov, A. A., Lyubetsky, V. A. & Gelfand, M. S. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA 14, 717–735 (2008).
Kalvari, I. et al. Non-coding RNA analysis using the Rfam database. Curr. Protoc. Bioinformatics. 62, e51 (2018).
Henkin, T. M., Glass, B. L. & Grundy, F. J. Analysis of the Bacillus subtilis tyrS gene: conservation of a regulatory sequence in multiple tRNA synthetase genes. J. Bacteriol. 174, 1299–1306 (1992).
Grundy, F. J., Yousef, M. R. & Henkin, T. M. Monitoring uncharged tRNA during transcription of the Bacillus subtilis glyQS gene. J. Mol. Biol. 346, 73–81 (2005).
Murakami, H., Ohta, A., Goto, Y., Sako, Y. & Suga, H. Flexizyme as a versatile tRNA acylation catalyst and the application for translation. Nucleic Acids Symp. Ser. (Oxf.) 50, 35–36 (2006).
Golden, B. L., Chen, J. & Luptak, A. Ribozyme with tRNA synthetase activity and methods of manufacturing and using the same. US patent 20,160,348,108 (2016).
Ke, A. & Doudna, J. A. Crystallization of RNA and RNA–protein complexes. Methods 34, 408–414 (2004).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010).
Pape, T. et al. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Crystallogr. 37, 843–844 (2004).
Sheldrick, G. M. et al. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D 66, 479–485 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Emsley, P. et al. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Guérout-Fleury, A.-M., Frandsen, N. & Stragier, P. Plasmids for ectopic integration in Bacillus subtilis. Gene 180, 57–61 (1996).
Anagnostopoulos, C. & Spizizen, J. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81, 741–746 (1961).
Chen, L., James, L. P. & Helmann, J. D. Metalloregulation in Bacillus subtilis: isolation and characterization of two genes differentially repressed by metal ions. J. Bacteriol. 175, 5428–5437 (1993).
Miller, J. H. Experiments in Molecular Genetics (Cold Spring Harbor Laboratory, 1972).
Acknowledgements
The authors thank A.J. Sachla and J.D. Helmann for advice on the β-galactosidase assay, the Bacillus Genetic Stock Center, the Nicholson Lab for the use of their spectrophotometer and the Fromme Lab for the use of their microscope. This work is supported by the National Institutes of Health (NIH) (grant nos. GM118174 and GM116632 to A.K.). This work is based on research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the NIH (grant no. P30 GM124165). The Pilatus 6 M detector on the 24-ID-C beamline is funded by a NIH-ORIP HEI grant (no. S10 RR029205). This research used the resources of the Advanced Photon Source, a US Department of Energy Office of Science User Facility operated for the US Department of Energy Office of Science by Argonne National Laboratory (contract no. DE-AC02-06CH11357).
Author information
Authors and Affiliations
Contributions
R.A.B., J.C.G. and A.K. designed the research. R.A.B. was the main contributor to the structure-function analysis. J.C.G. contributed to experimental design and structural refinement. R.A.B. and A.K. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Anke Sparmann was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data 1 Size exclusion chromatographic analysis and mutagenesis guide.
a, Size exclusion chromatogram of Mtb-ileS folded with (blue) and without (orange) tRNAIle. b, Urea denaturing PAGE gel of peak fractions from T-box + tRNA peak. c, Secondary structure model of tyrS T-box from Bacillus subtilis (Bsub-tyrS). Boxes highlight location and identity of each mutation. d, Conversion table for residue notation in Mtb-ileS and Bsub-tyrS. Fold difference from WT is the difference between the average of three induction measurements at 1 hour for WT versus mutation of indicated residue.
Extended Data 2 Details of AntiS-tRNA and AntiS-Stem-III interactions.
a, Detail of G132-U160 wobble pair in the aminoacylation sensing pocket showing AntiS (cyan), tRNA (pale blue) and linker (yellow). Hydrogen bonds represented by black dashes. Magnesium ion represented by green sphere. b, View of AntiS bound to tRNA showing tRNA 3′-end exposed in a large opening. c, Stem-III (pink) and AntiS minor groove interactions. d, Details of G98 interactions with AntiS. e, Details of A118 type-I A-minor interaction with AntiS.
Extended Data 3 Structural details and size exclusion chromatography of Mtb-ileS with tRNAIle-cP.
a, Tertiary structure model of Mtb-ileS in complex with tRNAIle-cP. b, Detail of AntiS interaction with tRNAIle-cP NCCA sequence. Hydrogen bonds represented by black dashes. Asterisk indicates residues mutated from wildtype sequence. c, Size exclusion chromatograms of Mtb-ileS folded with tRNAIle-cP (blue) and with tRNAIle-OH (orange). d, Urea denaturing PAGE gel of peak fractions from T-box + tRNAIle-cP peak.
Extended Data 4 Mechanistic diagram of atypical T-box translational regulation.
a, tRNA recruitment and decoding. b, Pseudoknot formation positions AntiS to interact with tRNA NCCA sequence. c, Transient intermediate where aminoacylation sensing module interacts with uncharged (top) and charged (bottom) tRNA. d, Favorable interactions between uncharged tRNA and linker locks T-box into ON conformation. Exposed Shine–Dalgarno (SDS) allows translation initiation. e, Steric clashing between charged tRNA and linker unravels the aminoacylation sensing module leading to alternative base pairing. Sequestrator formation prevents ribosome access to SDS and prevents translation.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2
Supplementary Table 3
Source data for Figs. 2b and 3f
Rights and permissions
About this article
Cite this article
Battaglia, R.A., Grigg, J.C. & Ke, A. Structural basis for tRNA decoding and aminoacylation sensing by T-box riboregulators. Nat Struct Mol Biol 26, 1106–1113 (2019). https://doi.org/10.1038/s41594-019-0327-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-019-0327-6
This article is cited by
-
Direct observation of tRNA-chaperoned folding of a dynamic mRNA ensemble
Nature Communications (2023)
-
Structural and dynamic mechanisms for coupled folding and tRNA recognition of a translational T-box riboswitch
Nature Communications (2023)
-
Incorporation of nonstandard amino acids into proteins: principles and applications
World Journal of Microbiology and Biotechnology (2020)
-
Engineering the Translational Machinery for Biotechnology Applications
Molecular Biotechnology (2020)
-
T-box RNA gets boxed
Nature Structural & Molecular Biology (2019)