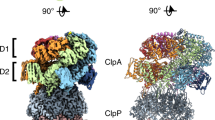
Abstract
The ClpXP machinery is a two-component protease complex that performs targeted protein degradation in bacteria and mitochondria. The complex consists of the AAA+ chaperone ClpX and the peptidase ClpP. The hexameric ClpX utilizes the energy of ATP binding and hydrolysis to engage, unfold and translocate substrates into the catalytic chamber of tetradecameric ClpP, where they are degraded. Formation of the complex involves a symmetry mismatch, because hexameric AAA+ rings bind axially to the opposing stacked heptameric rings of the tetradecameric ClpP. Here we present the cryo-EM structure of ClpXP from Listeria monocytogenes. We unravel the heptamer-hexamer binding interface and provide novel insight into the ClpX-ClpP cross-talk and activation mechanism. Comparison with available crystal structures of ClpP and ClpX in different states allows us to understand important aspects of the complex mode of action of ClpXP and provides a structural framework for future pharmacological applications.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The cryo-EM map of LmClpXP1−2 has been deposited at the EMDB with the accession code EMD-10162. The corresponding molecular models of LmClpX and LmClpP1−2 have been deposited at the wwPDB with accession codes PDB 6SFW and PDB 6SFX, respectively. Source data for Fig. 4, Fig. 5b,c, Supplementary Fig. 1a and Supplementary Fig. 6 are available online. All data used in this study are available from the corresponding authors upon reasonable request.
References
Bhandari, V. et al. The role of ClpP protease in bacterial pathogenesis and human diseases. ACS Chem. Biol. 13, 1413–1425 (2018).
Gaillot, O., Pellegrini, E., Bregenholt, S., Nair, S. & Berche, P. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 35, 1286–1294 (2000).
Yu, A. Y. H. & Houry, W. A. ClpP: a distinctive family of cylindrical energy-dependent serine proteases. FEBS Lett. 581, 3749–3757 (2007).
Brötz-Oesterhelt, H. et al. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat. Med. 11, 1082–1087 (2005).
Sauer, R. T. & Baker, T. A. AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 80, 587–612 (2011).
Beuron, F. et al. At sixes and sevens: characterization of the symmetry mismatch of the ClpAP chaperone-assisted protease. J. Struct. Biol. 123, 248–259 (1998).
Grimaud, R., Kessel, M., Beuron, F., Steven, A. C. & Maurizi, M. R. Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J. Biol. Chem. 273, 12476–12481 (1998).
Wang, J., Hartling, J. A. & Flanagan, J. M. The structure of ClpP at 2.3 A resolution suggests a model for ATP-dependent proteolysis. Cell 91, 447–456 (1997).
Banecki, B., Wawrzynow, A., Puzewicz, J., Georgopoulos, C. & Zylicz, M. Structure-function analysis of the zinc-binding region of the Clpx molecular chaperone. J. Biol. Chem. 276, 18843–18848 (2001).
Baker, T. A. & Sauer, R. T. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim. Biophys. Acta 1823, 15–28 (2012).
Bewley, M. C., Graziano, V., Griffin, K. & Flanagan, J. M. The asymmetry in the mature amino-terminus of ClpP facilitates a local symmetry match in ClpAP and ClpXP complexes. J. Struct. Biol. 153, 113–128 (2006).
Joshi, S. A., Hersch, G. L., Baker, T. A. & Sauer, R. T. Communication between ClpX and ClpP during substrate processing and degradation. Nat. Struct. Mol. Biol. 11, 404–411 (2004).
Martin, A., Baker, T. A. & Sauer, R. T. Distinct static and dynamic interactions control ATPase-peptidase communication in a AAA+ protease. Mol. Cell 27, 41–52 (2007).
Kim, Y. I. et al. Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nat. Struct. Biol. 8, 230–233 (2001).
Gersch, M. et al. AAA+ chaperones and acyldepsipeptides activate the ClpP protease via conformational control. Nat. Commun. 6, 6320 (2015).
Kirstein, J. et al. The antibiotic ADEP reprogrammes ClpP, switching it from a regulated to an uncontrolled protease. EMBO Mol. Med 1, 37–49 (2009).
Schmitz, K. R., Carney, D. W., Sello, J. K. & Sauer, R. T. Crystal structure of Mycobacterium tuberculosis ClpP1P2 suggests a model for peptidase activation by AAA+ partner binding and substrate delivery. Proc. Natl Acad. Sci. USA 111, E4587–E4595 (2014).
Alexopoulos, J. et al. Structural determinants stabilizing the axial channel of ClpP for substrate translocation. Mol. Microbiol. 90, 167–180 (2013).
Zeiler, E. et al. Vibralactone as a tool to study the activity and structure of the ClpP1P2 complex from Listeria monocytogenes. Angew. Chem. Int. Ed. Engl. 50, 11001–11004 (2011).
Balogh, D. et al. Insights into ClpXP proteolysis: heterooligomerization and partial deactivation enhance chaperone affinity and substrate turnover in Listeria monocytogenes. Chem. Sci. 8, 1592–1600 (2017).
Amor, A. J., Schmitz, K. R., Sello, J. K., Baker, T. A. & Sauer, R. T. Highly dynamic interactions maintain kinetic stability of the ClpXP protease during the ATP-fueled mechanical cycle. ACS Chem. Biol. 11, 1552–1560 (2016).
Hersch, G. L., Burton, R. E., Bolon, D. N., Baker, T. A. & Sauer, R. T. Asymmetric interactions of ATP with the AAA+ ClpX6 unfoldase: allosteric control of a protein machine. Cell 121, 1017–1027 (2005).
Ortega, J., Singh, S. K., Ishikawa, T., Maurizi, M. R. & Steven, A. C. Visualization of substrate binding and translocation by the ATP-dependent protease, ClpXP. Mol. Cell 6, 1515–1521 (2000).
Wagner, T. et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2019, 218 (2019).
Moriya, T. et al. High-resolution single particle analysis from electron cryo-microscopy images using SPHIRE. J. Vis. Exp. https://doi.org/10.3791/55448 (2017).
Donaldson, L. W., Wojtyra, U. & Houry, W. A. Solution structure of the dimeric zinc binding domain of the chaperone ClpX. J. Biol. Chem. 278, 48991–48996 (2003).
Wojtyra, U. A., Thibault, G., Tuite, A. & Houry, W. A. The N-terminal zinc binding domain of ClpX is a dimerization domain that modulates the chaperone function. J. Biol. Chem. 278, 48981–48990 (2003).
Stinson, B. M. et al. Nucleotide binding and conformational switching in the hexameric ring of a AAA+ machine. Cell 153, 628–639 (2013).
Trabuco, L. G., Villa, E., Mitra, K., Frank, J. & Schulten, K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16, 673–683 (2008).
Fux, A., Korotkov, V. S., Schneider, M., Antes, I. & Sieber, S. A. Chemical cross-linking enables drafting ClpXP Proximity maps and taking snapshots of insitu interaction networks. Cell Chem. Biol. 26, 48–59.e7 (2019).
Leodolter, J., Warweg, J. & Weber-Ban, E. The mycobacterium tuberculosis ClpP1P2 protease interacts asymmetrically with Its ATPase partners ClpX and ClpC1. PLOS One. 10, e0125345 (2015).
Dahmen, M., Vielberg, M.-T., Groll, M. & Sieber, S. A. Structure and mechanism of the caseinolytic protease ClpP1/2 heterocomplex from Listeria monocytogenes. Angew. Chem. Int. Ed. Engl. 54, 3598–3602 (2015).
Glynn, S. E., Martin, A., Nager, A. R., Baker, T. A. & Sauer, R. T. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell 139, 744–756 (2009).
Gates, S. N. et al. Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science 357, 273–279 (2017).
Lo, Y.-H. et al. Cryo-EM structure of the essential ribosome assembly AAA-ATPase Rix7. Nat. Commun. 10, 513 (2019).
Ripstein, Z. A., Huang, R., Augustyniak, R., Kay, L. E. & Rubinstein, J. L. Structure of a AAA+ unfoldase in the process of unfolding substrate. eLife 6, 43 (2017).
Gatsogiannis, C. et al. Tc toxin activation requires unfolding and refolding of a β-propeller. Nature 563, 209–213 (2018).
la Peña, de et al. 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science 362, eaav0725 (2018).
Majumder, P. et al. Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle. Proc. Natl Acad. Sci. USA 116, 534–539 (2019).
Martin, A., Baker, T. A. & Sauer, R. T. Diverse pore loops of the AAA+ ClpX machine mediate unassisted and adaptor-dependent recognition of ssrA-tagged substrates. Mol. Cell 29, 441–450 (2008).
Lee, B.-G. et al. Structures of ClpP in complex with acyldepsipeptide antibiotics reveal its activation mechanism. Nat. Publ. Group 17, 471–478 (2010).
Li, D. H. S. et al. Acyldepsipeptide antibiotics induce the formation of a structured axial channel in ClpP: A model for the ClpX/ClpA-bound state of ClpP. Chem. Biol. 17, 959–969 (2010).
Jennings, L. D., Bohon, J., Chance, M. R. & Licht, S. The ClpP N-terminus coordinates substrate access with protease active site reactivity. Biochemistry 47, 11031–11040 (2008).
Geiger, S. R., Böttcher, T., Sieber, S. A. & Cramer, P. A conformational switch underlies ClpP protease function. Angew. Chem. Int. Ed. Engl. 50, 5749–5752 (2011).
Gersch, M., List, A., Groll, M. & Sieber, S. A. Insights into structural network responsible for oligomerization and activity of bacterial virulence regulator caseinolytic protease P (ClpP) protein. J. Biol. Chem. 287, 9484–9494 (2012).
Kimber, M. S. et al. Structural and theoretical studies indicate that the cylindrical protease ClpP samples extended and compact conformations. Structure 18, 798–808 (2010).
Ye, F. et al. Helix unfolding/refolding characterizes the functional dynamics of Staphylococcus aureus Clp protease. J. Biol. Chem. 288, 17643–17653 (2013).
Zhang, J. et al. Structural switching of Staphylococcus aureus Clp protease: a key to understanding protease dynamics. J. Biol. Chem. 286, 37590–37601 (2011).
Ni, T. et al. Characterization of Gain-of-function mutant provides new insights into ClpP structure. ACS Chem. Biol. 11, 1964–1972 (2016).
Stahl, M. & Sieber, S. A. An amino acid domino effect orchestrates ClpP’s conformational states. Curr. Opin. Chem. Biol. 40, 102–110 (2017).
Böttcher, T. & Sieber, S. A. Beta-lactones as privileged structures for the active-site labeling of versatile bacterial enzyme classes. Angew. Chem. Int. Ed. Engl. 47, 4600–4603 (2008).
Frees, D. et al. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 54, 1445–1462 (2004).
Cole, A. et al. Inhibition of the mitochondrial protease ClpP as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell 27, 864–876 (2015).
Eisele, M. R. et al. Expanded coverage of the 26S proteasome conformational landscape reveals mechanisms of peptidase gating. Cell Rep. 24, 1301–1315.e5 (2018).
Smith, D. M. et al. Docking of the proteasomal ATPases‘ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol. Cell 27, 731–744 (2007).
Wehmer, M. et al. Structural insights into the functional cycle of the ATPase module of the 26S proteasome. Proc. Natl Acad. Sci. USA 114, 1305–1310 (2017).
Deville, C., Franke, K., Mogk, A., Bukau, B. & Saibil, H. R. Two-step activation mechanism of the ClpB disaggregase for sequential substrate threading by the main ATPase motor. Cell Rep. 27, 3433–3446.e4 (2019).
Liu, H. & Naismith, J. H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91 (2008).
Gatsogiannis, C. et al. Membrane insertion of a Tc toxin in near-atomic detail. Nat. Struct. Mol. Biol. 23, 884–890 (2016).
Grant, T. & Grigorieff, N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife 4, e06980 (2015).
Penczek, P. A. et al. CTER-rapid estimation of CTF parameters with error assessment. Ultramicroscopy 140, 9–19 (2014).
Tang, G. et al. EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Wagner, T. et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218 (2019).
Tanaka, Y. et al. Cryo-EM reveals the asymmetric assembly of squid hemocyanin. IUCrJ 6, 426–437 (2019).
Gatsogiannis, C. et al. Tc toxin activation requires unfolding and refolding of a β-propeller. Nature 563, 209−213 (2018).
Terwilliger, T. C., Sobolev, O. V., Afonine, P. V. & Adams, P. D. Automated map sharpening by maximization of detail and connectivity. Acta Crystallogr D. Struct. Biol. 74, 545–559 (2018).
Biasini, M. et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 (2014).
Pettersen, E. F. et al. UCSF Chimera?A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Frenz, B., Walls, A. C., Egelman, E. H., Veesler, D. & DiMaio, F. RosettaES: a sampling strategy enabling automated interpretation of difficult cryo-EM maps. Nat. Methods 14, 797–800 (2017).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Singharoy, A. et al. Molecular dynamics-based refinement and validation for sub-5 Å cryo-electron microscopy maps. eLife 5, 213 (2016).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D Struct. Biol. 74, 519–530 (2018).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, (213–221 (2010).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph 14, –33−38 (1996).
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005).
Huang, J. et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Barad, B. A. et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015).
Ashkenazy, H., Erez, E., Martz, E., Pupko, T. & Ben-Tal, N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533 (2010).
Masood, T. B., Sandhya, S., Chandra, N. & Natarajan, V. CHEXVIS: a tool for molecular channel extraction and visualization. BMC Bioinformatics 16, 119 (2015).
Goddard, T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Englander, S. W. & Kallenbach, N. R. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q. Rev. Biophys. 16, 521–655 (1983).
Wales, T. E. & Engen, J. R. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom. Rev. 25, 158–170 (2006).
Acknowledgements
We thank O. Hofnagel for assistance in electron microscopy and Dr. M. Lakemeyer for building ClpX homology models. We are grateful to Dr. M. Haslbeck, G. M. Feind and F. Rührnößl for HDX-MS measurements. This work was supported by the Max Planck Society (to S.R.), the European Research Council (FP7/2007-2013) (grant no. 615984) (to S.R.) and the Deutsche Forschungsgemeinschaft (SFB1035) (to S.A.S).
Author information
Authors and Affiliations
Contributions
S.A.S. and S.R. designed the study. C.G. screened and optimized samples, prepared cryo-EM grids and processed and analyzed cryo-EM data. D.B. cloned, overexpressed and purified proteins, optimized sample preparation, conducted activity assays and gel filtration measurements and analyzed HDX-MS data. C.G. and F.M. built atomic models. C.G. and D.B. prepared figures, C.G., D.B., S.A.S. and S.R. wrote the manuscript. All authors discussed the results.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Ines Chen was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 EM analysis of the ClpXP1–2 dimer.
a) Size exclusion chromatography of ClpX and ClpP1–2 mixtures on a Superose 6 increase 10/300 column. For the EM studies, a sample at 12 mL was taken. Note that the (ClpXP1–2)2 peak is absent with ClpXΔZBD. Source data are available online. b) SDS-PAGE of the isolated (ClpXP1–2)2 complex. c–d) Subarea of a negative stain EM micrograph of the isolated (ClpXP1–2)2 prior (c) and after (d) crosslinking; Scale bar, 200 nm e) Fourier Shell Correlation (FSC) between two independently refined half maps f) Low resolution cryo-EM density of the ClpXP1–2-dimer g) 3D clustering of the ClpXP1–2-dimer dataset.
Supplementary Figure 2 Cryo-EM analysis of ClpXP1–2.
a) Subarea of a typical low-dose cryo-EM micrograph of ClpXP1–2. ClpXP1–2 particles were selected and extracted from ClpXP1–2-ClpXP1–2 dimers using crYOLO and highlighted in red boxes. Scale bar, 100 nm b) Representative reference-free 2D class averages of ClpXP1–2. Scale bar, 25 nm c) Fourier Shell Correlation (FSC) between two independently refined half maps d) Side and cut-off view of the density map colored according to the local resolution e) Orientation distribution of the particles used in the final refinement round f-g) Superposition of segments of the molecular model of ClpP (f) and ClpX (g) with the cryo-EM density (transparent surface).
Supplementary Figure 3 ClpP-bound ClpX subunits adopt a nucleotide-loadable conformation.
Molecular models of the six ClpX subunits are shown as ribbon diagrams, with the large domain highlighted in orange and the small domain in red. Note the high similarity between the ClpX subunits, except the arrangement of their IGF-loops. The gray arrow indicates the IGF-loop of subunit Q that adopts an ‘extended’ conformation. The inset shows nucleotide-loadable (L; upper image) and unloadable (U; lower image) subunits of ATPΥS-bound EcClpX (PDB 4I81). Structural comparison of the six ClpX subunits with the L and U subunit of EcClpX (note the respective RMSD values of the Cα atoms) indicate that all subunits of ClpP-bound ClpX adopt a loadable conformation.
Supplementary Figure 4 HDX-MS of the ClpXP1–2 complex.
Changes in deuterium uptake after complex formation are mapped on the amino acid sequence of a) ClpX, b) ClpP1 and c) ClpP2 for the respective exposure times. Increased deuterium uptake upon complex formation is shown in red, decreased deuterium uptake is depicted in blue. Dark gray represents no coverage. Please refer to the Methods section for the calculation of the relative deuterium uptake values. Averages of two independent measurements are shown.
Supplementary Figure 5 Cryo-EM density for the IGF-loops interfaces and the ClpP2 catalytic active site.
a) Densities for the six IGF-loops interactions are shown with the corresponding atomic models. ClpX and ClpP2 densities are shown as gray and green transparent isosurface, respectively. b) Superposition of the catalytic residues S98 (S98A), H123 and D172 in ClpX-bound LmClpP1-S98A/P2-S98A (cryo-EM) (extended active state) and SaClpP (compact inactive state) (PDB 4EMM), shown with the cryo-EM density. The catalytic residues of ClpX-bound LmClpP1–P2 adopt the active conformation.
Supplementary Figure 6 Activity assays of ClpX and ClpP2 mutants of the IGF-loop/hydrophobic groove interface.
a) Peptidase activity of ClpP1–2 with respective ClpP2 mutants (0.71 μM (ClpP1–2)14, 100 μM Ac-Ala-hArg-2-Aoc-ACC). b) ATPase activity of ClpX mutants (0.33 μM ClpX6, 20 mM ATP). c) Protease activity of ClpXP1–2 with ClpP2 and ClpX mutants (0.2 μM (ClpP1–2)14, 0.4 μM ClpX6, 0.8 μM GFP-SsrA). Data are normalized to the wild type as 100% (n = 6, black lines denote means). Source data for graphs in a-c are available online. d) Mapping of the ClpP2 and ClpX mutations on the protein structure. Mutation sites are shown with red sticks.
Supplementary Figure 7 The N-terminal loops of ClpX-bound ClpP2 subunits adopt the “up” conformation.
Cryo-EM density map (mesh) with the molecular model highlighting the N-terminal domain of the seven ClpP subunits. Residues 8-17 are not resolved, but the fragmented cryo-EM density indicates that all flexible N-terminal loops adopt the “up” conformation (indicated by dashed lines). For better comparison, the molecular model of a subunit of EcClpP (PDB 1YG6) with the N-terminus in the “down” conformation (orange) is also shown.
Supplementary Figure 8 Possible interactions between pore-2 loops of ClpX with the N-termini of ClpP.
a-f) Cryo-EM density map with the molecular model, highlighting the interaction area between the pore-2 loops of ClpX and the N-termini of ClpP. The six pore-2 loops of ClpX and residues 7-16 of the N-termini of seven subunits of ClpP2 are not resolved. Possible arrangements of these regions are indicated by dashed lines, based on their anchor points and number of residues. Note that the pore-2 loops of chains Q and P point into a cleft formed by three ClpP N-termini (b,c). This topological analysis also suggests that the pore-2 loops of chains O and T (a,f) do not show any interactions with the N-termini of ClpP. However, an unusual stretched conformation of these pore-2 loops towards ClpP cannot be excluded. Pore-2 loop of chain S is positioned in direct proximity to the N-terminus of chain N (c) whereas the pore-2-loop of chain R is positioned between two ClpP N-termini (M and N) (e).
Supplementary Figure 9 Alignment of ClpP sequences.
Mt = Mycobacterium tuberculosis (ClpP2), Pa = Pseudomonas aeruginosa (ClpP1), Cd = Clostridium difficile (ClpP1 and ClpP2), Ec = Escherichia coli, Lm = Listeria monocytogenes (ClpP2), Bs = Bacillus subtilis, Sa = Staphylococcus aureus.
Supplementary information
Supplementary Information
Supplementary Figs. 1–9 and Table 1
Supplementary Video 1
Flexibility of ClpXP1/2 dimers in 2D.
Supplementary Video 2
Structural comparison between ClpX-bound and ADEP-bound ClpP. The video shows a simple linear interpolation between ClpX-bound ClpP1/2 (cryo-EM) (extended active conformation) and the available crystal structure of B. subtilis ADEP2-bound ClpP (PDB 3KTK41) (extended active open conformation), first along the ClpP1 and finally along the ClpP2 face (see Fig. 6c). Note the widening of the pore in the ADEP-bound structure.
Rights and permissions
About this article
Cite this article
Gatsogiannis, C., Balogh, D., Merino, F. et al. Cryo-EM structure of the ClpXP protein degradation machinery. Nat Struct Mol Biol 26, 946–954 (2019). https://doi.org/10.1038/s41594-019-0304-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-019-0304-0
This article is cited by
-
Insights into the structure-function relationship of the NorQ/NorD chaperones from Paracoccus denitrificans reveal shared principles of interacting MoxR AAA+/VWA domain proteins
BMC Biology (2023)
-
A closed translocation channel in the substrate-free AAA+ ClpXP protease diminishes rogue degradation
Nature Communications (2023)
-
Structural basis of impaired disaggregase function in the oxidation-sensitive SKD3 mutant causing 3-methylglutaconic aciduria
Nature Communications (2023)
-
Structural basis of prokaryotic ubiquitin-like protein engagement and translocation by the mycobacterial Mpa-proteasome complex
Nature Communications (2022)
-
Quality control of the mitochondrial proteome
Nature Reviews Molecular Cell Biology (2021)