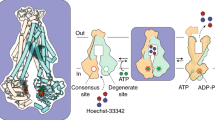
Abstract
Much structural information has been amassed on ATP-binding cassette (ABC) transporters, including hundreds of structures of isolated domains and an increasing array of full-length transporters. The structures capture different steps in the transport cycle and have aided in the design and interpretation of computational simulations and biophysics experiments. These data provide a maturing, although still incomplete, elucidation of the protein dynamics and mechanisms of substrate selection and transit through the transporters. We present an updated view of the classical alternating-access mechanism as it applies to eukaryotic ABC transporters, focusing on type I exporters. Our model helps frame the progress in, and remaining questions about, transporter energetics, how substrates are selected and how ATP is consumed to perform work at the molecular scale. Many human ABC transporters are associated with disease; we highlight progress in understanding their pharmacology through the lens of structural biology and describe how this knowledge suggests approaches to pharmacologically targeting these transporters.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ford, R. C. & Beis, K. Learning the ABCs one at a time: structure and mechanism of ABC transporters. Biochem. Soc. Trans. 47, 23–36 (2019).
Cui, J. & Davidson, A. L. ABC solute importers in bacteria. Essays Biochem. 50, 85–99 (2011).
Davidson, A. L., Dassa, E., Orelle, C. & Chen, J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72, 317–64 (2008).
Decottignies, A. & Goffeau, A. Complete inventory of the yeast ABC proteins. Nat. Genet. 15, 137–145 (1997).
Vasiliou, V., Vasiliou, K. & Nebert, D. W. Human ATP-binding cassette (ABC) transporter family. Hum. Genom. 3, 281–290 (2009).
Garcia, O., Bouige, P., Forestier, C. & Dassa, E. Inventory and comparative analysis of rice and Arabidopsis ATP-binding cassette (ABC) systems. J. Mol. Biol. 343, 249–265 (2004).
Quazi, F., Lenevich, S. & Molday, R. S. ABCA4 is an N-retinylidene-phosphatidylethanolamine and phosphatidylethanolamine importer. Nat. Commun. 3, 925 (2012).
Xiong, J., Feng, J., Yuan, D., Zhou, J. & Miao, W. Tracing the structural evolution of eukaryotic ATP binding cassette transporter superfamily. Sci. Rep. 5, 16724 (2015).
Theodoulou, F. L. & Kerr, I. D. ABC transporter research: going strong 40 years on. Biochem. Soc. Trans. 43, 1033–1040 (2015).
Riordan, J. R. et al. Amplification of P-glycoprotein genes in multidrug-resistant mammalian cell lines. Nature 316, 817–819 (1985).
Robey, R. W. et al. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer 18, 452–464 (2018).
Cavalheiro, M., Pais, P., Galocha, M. & Teixeira, M. C. Host-pathogen interactions mediated by MDR transporters in fungi: as pleiotropic as it gets! Genes (Basel) 9, E332 (2018).
Du, D. et al. Multidrug efflux pumps: structure, function and regulation. Nat. Rev. Microbiol. 16, 523–539 (2018).
Moitra, K. & Dean, M. Evolution of ABC transporters by gene duplication and their role in human disease. Biol. Chem. 392, 29–37 (2011).
Gaudet, R. & Wiley, D. C. Structure of the ABC ATPase domain of human TAP1, the transporter associated with antigen processing. EMBO J. 20, 4964–4972 (2001).
Hung, L. W. et al. Crystal structure of the ATP-binding subunit of an ABC transporter. Nature 396, 703–707 (1998).
Chen, J., Lu, G., Lin, J., Davidson, A. L. & Quiocho, F. A. A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol. Cell 12, 651–661 (2003).
Scapin, G., Potter, C. S. & Carragher, B. Cryo-EM for small molecules discovery, design, understanding, and application. Cell Chem. Biol. 25, 1318–1325 (2018).
Frank, J. Advances in the field of single-particle cryo-electron microscopy over the last decade. Nat. Protoc. 12, 209–212 (2017).
Jardetzky, O. Simple allosteric model for membrane pumps. Nature 211, 969–970 (1966).
Wen, P. C., Verhalen, B., Wilkens, S., Mchaourab, H. S. & Tajkhorshid, E. On the origin of large flexibility of P-glycoprotein in the inward-facing state. J. Biol. Chem. 288, 19211–19220 (2013).
Jin, M. S., Oldham, M. L., Zhang, Q. & Chen, J. Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature 490, 566–569 (2012).
Li, J., Jaimes, K. F. & Aller, S. G. Refined structures of mouse P-glycoprotein. Protein Sci. 23, 34–46 (2014).
Lee, J. Y., Yang, J. G., Zhitnitsky, D., Lewinson, O. & Rees, D. C. Structural basis for heavy metal detoxification by an Atm1-type ABC exporter. Science 343, 1133–1136 (2014).
Esser, L. et al. Structures of the multidrug transporter P-glycoprotein reveal asymmetric ATP binding and the mechanism of polyspecificity. J. Biol. Chem. 292, 446–461 (2017).
Alam, A. et al. Structure of a zosuquidar and UIC2-bound human-mouse chimeric ABCB1. Proc. Natl Acad. Sci. USA 115, E1973–E1982 (2018).
Johnson, Z. L. & Chen, J. ATP binding enables substrate release from multidrug resistance protein 1. Cell 172, 81–89.e10 (2018).
Dawson, R. J. & Locher, K. P. Structure of the multidrug ABC transporter Sav1866 from Staphylococcus aureus in complex with AMP-PNP. FEBS Lett. 581, 935–938 (2007).
Barth, K. et al. Conformational coupling and trans-inhibition in the human antigen transporter ortholog TmrAB resolved with dipolar EPR spectroscopy. J. Am. Chem. Soc. 140, 4527–4533 (2018).
Kim, Y. & Chen, J. Molecular structure of human P-glycoprotein in the ATP-bound, outward-facing conformation. Science 359, 915–919 (2018).
Hutter, C. A. J. et al. The extracellular gate shapes the energy profile of an ABC exporter. Nat. Commun. 10, 2260 (2019).
Kodan, A. et al. Structural basis for gating mechanisms of a eukaryotic P-glycoprotein homolog. Proc. Natl Acad. Sci. USA 111, 4049–4054 (2014).
Bountra, K. et al. Structural basis for antibacterial peptide self-immunity by the bacterial ABC transporter McjD. EMBO J. 36, 3062–3079 (2017).
Choudhury, H. G. et al. Structure of an antibacterial peptide ATP-binding cassette transporter in a novel outward occluded state. Proc. Natl Acad. Sci. USA 111, 9145–9150 (2014).
Alam, A., Kowal, J., Broude, E., Roninson, I. & Locher, K. P. Structural insight into substrate and inhibitor discrimination by human P-glycoprotein. Science 363, 753–756 (2019).
Loo, T. W., Bartlett, M. C. & Clarke, D. M. Drug binding in human P-glycoprotein causes conformational changes in both nucleotide-binding domains. J. Biol. Chem. 278, 1575–1578 (2003).
Johnson, Z. L. & Chen, J. Structural basis of substrate recognition by the multidrug resistance protein MRP1. Cell 168, 1075–1085.e9 (2017).
Penczek, P. A., Kimmel, M. & Spahn, C. M. Identifying conformational states of macromolecules by eigen-analysis of resampled cryo-EM images. Structure 19, 1582–1590 (2011).
Loveland, A. B. & Korostelev, A. A. Structural dynamics of protein S1 on the 70S ribosome visualized by ensemble cryo-EM. Methods 137, 55–66 (2018).
Frank, G. A. et al. Cryo-EM analysis of the conformational landscape of human P-glycoprotein (ABCB1) during its catalytic cycle. Mol. Pharmacol. 90, 35–41 (2016).
Harpole, T. J. & Delemotte, L. Conformational landscapes of membrane proteins delineated by enhanced sampling molecular dynamics simulations. Biochim. Biophys. Acta 1860, 909–926 (2018).
Marinelli, F. & Faraldo-Gómez, J. D. Ensemble-biased metadynamics: a molecular simulation method to sample experimental distributions. Biophys. J. 108, 2779–2782 (2015).
Lefèvre, F. & Boutry, M. Towards identification of the substrates of ATP-binding cassette transporters. Plant Physiol. 178, 18–39 (2018).
Dean, M., Rzhetsky, A. & Allikmets, R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 11, 1156–1166 (2001).
Cole, S. P. Multidrug resistance protein 1 (MRP1, ABCC1), a “multitasking” ATP-binding cassette (ABC) transporter. J. Biol. Chem. 289, 30880–30888 (2014).
Litman, T., Druley, T. E., Stein, W. D. & Bates, S. E. From MDR to MXR: new understanding of multidrug resistance systems, their properties and clinical significance. Cell. Mol. Life Sci. 58, 931–959 (2001).
Pedersen, J. M. et al. Substrate and method dependent inhibition of three ABC-transporters (MDR1, BCRP, and MRP2). Eur. J. Pharm. Sci. 103, 70–76 (2017).
Loo, T. W., Bartlett, M. C. & Clarke, D. M. Transmembrane segment 7 of human P-glycoprotein forms part of the drug-binding pocket. Biochem. J. 399, 351–359 (2006).
Loo, T. W., Bartlett, M. C. & Clarke, D. M. Simultaneous binding of two different drugs in the binding pocket of the human multidrug resistance P-glycoprotein. J. Biol. Chem. 278, 39706–39710 (2003).
Loo, T. W. & Clarke, D. M. Identification of residues within the drug-binding domain of the human multidrug resistance P-glycoprotein by cysteine-scanning mutagenesis and reaction with dibromobimane. J. Biol. Chem. 275, 39272–39278 (2000).
Loo, T. W. & Clarke, D. M. Thiol-reactive drug substrates of human P-glycoprotein label the same sites to activate ATPase activity in membranes or dodecyl maltoside detergent micelles. Biochem. Biophys. Res. Commun. 488, 573–577 (2017).
Aller, S. G. et al. Structure of P-glycoprotein reveals a molecular basis for poly specific drug binding. Science 323, 1718–1722 (2009).
Nicklisch, S. C. et al. Global marine pollutants inhibit P-glycoprotein: environmental levels, inhibitory effects, and cocrystal structure. Sci. Adv. 2, e1600001 (2016).
Oldham, M. L., Grigorieff, N. & Chen, J. Structure of the transporter associated with antigen processing trapped by herpes simplex virus. eLife 5, e21829 (2016).
Lehnert, E. & Tampé, R. Structure and dynamics of antigenic peptides in complex with TAP. Front. Immunol. 8, 10 (2017).
Koopmann, J. O., Post, M., Neefjes, J. J., Hämmerling, G. J. & Momburg, F. Translocation of long peptides by transporters associated with antigen processing (TAP). Eur. J. Immunol. 26, 1720–1728 (1996).
Ritz, U. et al. Impaired transporter associated with antigen processing (TAP) function attributable to a single amino acid alteration in the peptide TAP subunit TAP1. J. Immunol. 170, 941–946 (2003).
Armandola, E. A. et al. A point mutation in the human transporter associated with antigen processing (TAP2) alters the peptide transport specificity. Eur. J. Immunol. 26, 1748–1755 (1996).
Baldauf, C., Schrodt, S., Herget, M., Koch, J. & Tampé, R. Single residue within the antigen translocation complex TAP controls the epitope repertoire by stabilizing a receptive conformation. Proc. Natl Acad. Sci. USA 107, 9135–9140 (2010).
Geng, J., Pogozheva, I. D., Mosberg, H. I. & Raghavan, M. Use of functional polymorphisms to elucidate the peptide binding site of TAP complexes. J. Immunol. 195, 3436–3448 (2015).
Deverson, E. V. et al. Functional analysis by site-directed mutagenesis of the complex polymorphism in rat transporter associated with antigen processing. J. Immunol. 160, 2767–2779 (1998).
Lehnert, E. et al. Antigenic peptide recognition on the human ABC transporter TAP resolved by DNP-enhanced solid-state NMR spectroscopy. J. Am. Chem. Soc. 138, 13967–13974 (2016).
Nöll, A. et al. Crystal structure and mechanistic basis of a functional homolog of the antigen transporter TAP. Proc. Natl Acad. Sci. USA 114, E438–E447 (2017).
Mi, W. et al. Structural basis of MsbA-mediated lipopolysaccharide transport. Nature 549, 233–237 (2017).
Loo, T. W., Bartlett, M. C. & Clarke, D. M. Substrate-induced conformational changes in the transmembrane segments of human P-glycoprotein. Direct evidence for the substrate-induced fit mechanism for drug binding. J. Biol. Chem. 278, 13603–13606 (2003).
Spadaccini, R., Kaur, H., Becker-Baldus, J. & Glaubitz, C. The effect of drug binding on specific sites in transmembrane helices 4 and 6 of the ABC exporter MsbA studied by DNP-enhanced solid-state NMR. Biochim. Biophys. Acta 1860, 833–840 (2018).
Szewczyk, P. et al. Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein. Acta Crystallogr. D. Biol. Crystallogr. 71, 732–741 (2015).
Perez, C. et al. Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature 524, 433–438 (2015).
Szöllősi, D., Rose-Sperling, D., Hellmich, U. A. & Stockner, T. Comparison of mechanistic transport cycle models of ABC exporters. Biochim. Biophys. Acta 1860, 818–832 (2018).
Moeller, A. et al. Distinct conformational spectrum of homologous multidrug ABC transporters. Structure 23, 450–460 (2015).
Dastvan, R., Mishra, S., Peskova, Y. B., Nakamoto, R. K. & Mchaourab, H. S. Mechanism of allosteric modulation of P-glycoprotein by transport substrates and inhibitors. Science 364, 689–692 (2019).
Gu, R. X. et al. Conformational changes in the antibacterial peptide ATP binding cassette transporter McjD revealed by molecular dynamics simulations. Biochemistry 54, 5989–5998 (2015).
Grossmann, N. et al. Mechanistic determinants of the directionality and energetics of active export by a heterodimeric ABC transporter. Nat. Commun. 5, 5419 (2014).
Csanády, L., Vergani, P. & Gadsby, D. C. Structure, gating, and regulation of the Cftr anion channel. Physiol. Rev. 99, 707–738 (2019).
Pan, L. & Aller, S. G. Allosteric role of substrate occupancy toward the alignment of P-glycoprotein nucleotide binding domains. Sci. Rep. 8, 14643 (2018).
Procko, E., Ferrin-O’Connell, I., Ng, S. L. & Gaudet, R. Distinct structural and functional properties of the ATPase sites in an asymmetric ABC transporter. Mol. Cell 24, 51–62 (2006).
Mishra, S. et al. Conformational dynamics of the nucleotide binding domains and the power stroke of a heterodimeric ABC transporter. eLife 3, e02740 (2014).
Collauto, A., Mishra, S., Litvinov, A., McHaourab, H. S. & Goldfarb, D. Direct spectroscopic detection of ATP turnover reveals mechanistic divergence of ABC exporters. Structure 25, 1264–1274.e3 (2017).
Verhalen, B. et al. Energy transduction and alternating access of the mammalian ABC transporter P-glycoprotein. Nature 543, 738–741 (2017).
Borst, P. & Elferink, R. O. Mammalian ABC transporters in health and disease. Annu. Rev. Biochem. 71, 537–592 (2002).
Procko, E. & Gaudet, R. Antigen processing and presentation: TAPping into ABC transporters. Curr. Opin. Immunol. 21, 84–91 (2009).
Eggensperger, S. & Tampé, R. The transporter associated with antigen processing: a key player in adaptive immunity. Biol. Chem. 396, 1059–1072 (2015).
Palmeira, A., Sousa, E., Vasconcelos, M. H. & Pinto, M. M. Three decades of P-gp inhibitors: skimming through several generations and scaffolds. Curr. Med. Chem. 19, 1946–2025 (2012).
Ma, J. & Biggin, P. C. Substrate versus inhibitor dynamics of P-glycoprotein. Proteins 81, 1653–1668 (2013).
Dawson, R. J. & Locher, K. P. Structure of a bacterial multidrug ABC transporter. Nature 443, 180–185 (2006).
Perez, C. et al. Structural basis of inhibition of lipid-linked oligosaccharide flippase PglK by a conformational nanobody. Sci. Rep. 7, 46641 (2017).
Ho, H. et al. Structural basis for dual-mode inhibition of the ABC transporter MsbA. Nature 557, 196–201 (2018).
Praest, P., Liaci, A.M., Forster, F. & Wiertz, E. New insights into the structure of the MHC class I peptide-loading complex and mechanisms of TAP inhibition by viral immune evasion proteins. Mol. Immunol. https://doi.org/10.1016/j.molimm.2018.03.020 (2018).
Herbring, V., Bäucker, A., Trowitzsch, S. & Tampé, R. A dual inhibition mechanism of herpesviral ICP47 arresting a conformationally thermostable TAP complex. Sci. Rep. 6, 36907 (2016).
Oldham, M. L. et al. A mechanism of viral immune evasion revealed by cryo-EM analysis of the TAP transporter. Nature 529, 537–540 (2016).
Parreira, B. et al. Persistence of the ABCC6 genes and the emergence of the bony skeleton in vertebrates. Sci. Rep. 8, 6027 (2018).
Lee, J. Y. et al. Crystal structure of the human sterol transporter ABCG5/ABCG8. Nature 533, 561–564 (2016).
Taylor, N. M. I. et al. Structure of the human multidrug transporter ABCG2. Nature 546, 504–509 (2017).
Borghi, L., Kang, J., Ko, D., Lee, Y. & Martinoia, E. The role of ABCG-type ABC transporters in phytohormone transport. Biochem. Soc. Trans. 43, 924–930 (2015).
Luo, Y. L. et al. Tissue expression pattern of ABCG transporter indicates functional roles in reproduction of Toxocara canis. Parasitol. Res. 117, 775–782 (2018).
Manolaridis, I. et al. Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states. Nature 563, 426–430 (2018).
Jackson, S. M. et al. Structural basis of small-molecule inhibition of human multidrug transporter ABCG2. Nat. Struct. Mol. Biol. 25, 333–340 (2018).
Qian, H. et al. Structure of the human lipid exporter ABCA1. Cell 169, 1228–1239.e10 (2017).
Lee, J. Y. & Parks, J. S. ATP-binding cassette transporter AI and its role in HDL formation. Curr. Opin. Lipidol. 16, 19–25 (2005).
Smith, P. C. et al. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell 10, 139–149 (2002).
Kodan, A. et al. Inward- and outward-facing X-ray crystal structures of homodimeric P-glycoprotein CmABCB1. Nat. Commun. 10, 88 (2019).
Molinski, S. V. et al. Comprehensive mapping of cystic fibrosis mutations to CFTR protein identifies mutation clusters and molecular docking predicts corrector binding site. Proteins 86, 833–843 (2018).
de Wet, H. & Proks, P. Molecular action of sulphonylureas on KATP channels: a real partnership between drugs and nucleotides. Biochem. Soc. Trans. 43, 901–907 (2015).
Vergani, P., Lockless, S. W., Nairn, A. C. & Gadsby, D. C. CFTR channel opening by ATP-driven tight dimerization of its nucleotide-binding domains. Nature 433, 876–880 (2005).
Zhang, Z. & Chen, J. Atomic structure of the cystic fibrosis transmembrane conductance regulator. Cell 167, 1586–1597.e9 (2016).
Zhang, Z., Liu, F. & Chen, J. Conformational changes in CFTR upon phosphorylation and ATP binding. Cell 170, 483–491.e8 (2017).
Liu, F., Zhang, Z., Csanady, L., Gadsby, D. C. & Chen, J. Molecular structure of the human CFTR ion channel. Cell 169, 85–95.e8 (2017).
Zhang, Z., Liu, F. & Chen, J. Molecular structure of the ATP-bound, phosphorylated human CFTR. Proc. Natl Acad. Sci. USA 115, 12757–12762 (2018).
Tordai, H., Leveles, I. & Hegedűs, T. Molecular dynamics of the cryo-EM CFTR structure. Biochem. Biophys. Res. Commun. 491, 986–993 (2017).
Veit, G. et al. From CFTR biology toward combinatorial pharmacotherapy: expanded classification of cystic fibrosis mutations. Mol. Biol. Cell 27, 424–433 (2016).
Jih, K. Y. & Hwang, T. C. Vx-770 potentiates CFTR function by promoting decoupling between the gating cycle and ATP hydrolysis cycle. Proc. Natl Acad. Sci. USA 110, 4404–4409 (2013).
Yu, H. et al. Ivacaftor potentiation of multiple CFTR channels with gating mutations. J. Cyst. Fibros. 11, 237–245 (2012).
Li, N. et al. Structure of a pancreatic ATP-sensitive potassium channel. Cell 168, 101–110.e10 (2017).
Martin, G. M., Kandasamy, B., DiMaio, F., Yoshioka, C. & Shyng, S. L. Anti-diabetic drug binding site in a mammalian KATP channel revealed by Cryo-EM. eLife 6, e31054 (2017).
Martin, G. M. et al. Cryo-EM structure of the ATP-sensitive potassium channel illuminates mechanisms of assembly and gating. eLife 6, e24149 (2017).
Lee, K. P. K., Chen, J. & MacKinnon, R. Molecular structure of human KATP in complex with ATP and ADP. eLife 6, e32481 (2017).
Nichols, C. G. KATP channels as molecular sensors of cellular metabolism. Nature 440, 470–476 (2006).
Vedovato, N., Ashcroft, F. M. & Puljung, M. C. The nucleotide-binding sites of sur1: a mechanistic model. Biophys. J. 109, 2452–2460 (2015).
Acknowledgements
We thank A. Murray and members of the laboratories of R.G. and A. Murray for insightful discussions. This work was funded in part by NIH grant R01GM120996 (to R.G.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information: Katarzyna Marcinkiewicz was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Table 1
Rights and permissions
About this article
Cite this article
Srikant, S., Gaudet, R. Mechanics and pharmacology of substrate selection and transport by eukaryotic ABC exporters. Nat Struct Mol Biol 26, 792–801 (2019). https://doi.org/10.1038/s41594-019-0280-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-019-0280-4
This article is cited by
-
Identification and expression analysis of ATP-binding cassette (ABC) transporters revealed its role in regulating stress response in pear (Pyrus bretchneideri)
BMC Genomics (2024)
-
The cross talk of ubiquitination and chemotherapy tolerance in colorectal cancer
Journal of Cancer Research and Clinical Oncology (2024)
-
Advances in the structure, mechanism and targeting of chemoresistance-linked ABC transporters
Nature Reviews Cancer (2023)
-
Deep mutational scan of a drug efflux pump reveals its structure–function landscape
Nature Chemical Biology (2023)
-
On the interplay between lipids and asymmetric dynamics of an NBS degenerate ABC transporter
Communications Biology (2023)