Abstract
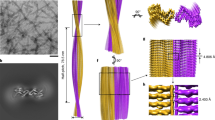
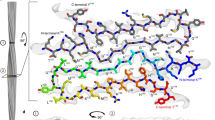
The atomic structure of the infectious, protease-resistant, β-sheet-rich and fibrillar mammalian prion remains unknown. Through the cryo-EM method MicroED, we reveal the sub-ångström-resolution structure of a protofibril formed by a wild-type segment from the β2–α2 loop of the bank vole prion protein. The structure of this protofibril reveals a stabilizing network of hydrogen bonds that link polar zippers within a sheet, producing motifs we have named ‘polar clasps’.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Shi, D., Nannenga, B. L., Iadanza, M. G. & Gonen, T.Three-dimensional electron crystallography of protein microcrystals. eLife 2, e01345 (2013).
de la Cruz, M. J. et al. Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED. Nat. Methods 14, 399–402 (2017).
Rodriguez, J. A., Eisenberg, D. S. & Gonen, T. Taking the measure of MicroED. Curr. Opin. Struct. Biol. 46, 79–86 (2017).
Rodriguez, J. A. et al. Structure of the toxic core of α-synuclein from invisible crystals. Nature 525, 486–490 (2015).
Krotee, P. et al. Atomic structures of fibrillar segments of hIAPP suggest tightly mated β-sheets are important for cytotoxicity. eLife 6, e19273 (2017).
Sawaya, M. R. et al. Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED. Proc. Natl. Acad. Sci. USA 113, 11232–11236 (2016).
Tuttle, M. D. et al.Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nat. Struct. Mol. Biol. 23, 409–415 (2016).
Schmidt, M. et al. Peptide dimer structure in an Aβ(1-42) fibril visualized with cryo-EM. Proc. Natl. Acad. Sci. USA 112, 11858–11863 (2015).
Fitzpatrick, A. W. P. et al. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 547, 185–190 (2017).
Wasmer, C. et al. Amyloid fibrils of the HET-s(218-289) prion form a β solenoid with a triangular hydrophobic core. Science 319, 1523–1526 (2008).
Rodriguez, J. A., Jiang, L. & Eisenberg, D. S. Toward the atomic structure of PrPSc. Cold Spring Harb. Perspect. Biol. 9, a031336 (2017).
Sawaya, M. R. et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature 447, 453–457 (2007).
Eisenberg, D. & Jucker, M.The amyloid state of proteins in human diseases. Cell 148, 1188–1203 (2012).
McKinley, M. P., Bolton, D. C. & Prusiner, S. B. A protease-resistant protein is a structural component of the scrapie prion. Cell 35, 57–62 (1983).
Kurt, T. D. & Sigurdson, C. J. Cross-species transmission of CWD prions. Prion 10, 83–91 (2016).
Goldschmidt, L., Teng, P. K., Riek, R. & Eisenberg, D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl. Acad. Sci. USA 107, 3487–3492 (2010).
Watts, J. C. et al. Evidence that bank vole PrP is a universal acceptor for prions. PLoS. Pathog. 10, e1003990 (2014).
Kurt, T. et al. The molecular basis for cross-species prion transmission. FASEB J. 30, 814.7 (2016).
Halfmann, R. et al. Opposing effects of glutamine and asparagine govern prion formation by intrinsically disordered proteins. Mol. Cell. 43, 72–84 (2011).
Zambrano, R. et al. PrionW: a server to identify proteins containing glutamine/asparagine rich prion-like domains and their amyloid cores. Nucleic Acids Res. 43, W331–W337 (2015).
Kurt, T. D. et al. Asparagine and glutamine ladders promote cross-species prion conversion. J. Biol. Chem. https://doi.org/10.1074/jbc.M117.794107 (2017).
Newberry, R. W. & Raines, R. T. A prevalent intraresidue hydrogen bond stabilizes proteins. Nat. Chem. Biol. 12, 1084–1088 (2016).
Nagle, J. F. & Morowitz, H. J. Molecular mechanisms for proton transport in membranes. Proc. Natl. Acad. Sci. USA 75, 298–302 (1978).
Yoder, M. D., Lietzke, S. E. & Jurnak, F. Unusual structural features in the parallel β-helix in pectate lyases. Structure 1, 241–251 (1993).
Perutz, M. F., Staden, R., Moens, L. & De Baere, I. Polar zippers. Curr. Biol. 3, 249–253 (1993).
Perutz, M. F., Johnson, T., Suzuki, M. & Finch, J. T. Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc. Natl. Acad. Sci. USA 91, 5355–5358 (1994).
Wiltzius, J. J. W. et al. Molecular mechanisms for protein-encoded inheritance. Nat. Struct. Mol. Biol. 16, 973–978 (2009).
Palatinus, L. et al. Hydrogen positions in single nanocrystals revealed by electron diffraction. Science 355, 166–169 (2017).
Hirano, Y., Takeda, K. & Miki, K. Charge-density analysis of an iron-sulfur protein at an ultra-high resolution of 0.48Å. Nature 534, 281–284 (2016).
Glaeser, R. M. & Downing, K. H. High-resolution electron crystallography of protein molecules. Ultramicroscopy 52, 478–486 (1993).
Zhong, S., Dadarlat, V. M., Glaeser, R. M., Head-Gordon, T. & Downing, K. H. Modeling chemical bonding effects for protein electron crystallography: the transferable fragmental electrostatic potential (TFESP) method. Acta Crystallogr. A 58, 162–170 (2002).
Jelsch, C. et al. Accurate protein crystallography at ultra-high resolution: valence electron distribution in crambin. Proc. Natl. Acad. Sci. USA 97, 3171–3176 (2000).
Zuo, J. M., Kim, M., O’Keeffe, M. & Spence, J. C. H. Direct observation of d-orbital holes and Cu–Cu bonding in Cu2O. Nature 401, 49–52 (1999).
Shi, D. et al. The collection of MicroED data for macromolecular crystallography. Nat. Protoc. 11, 895–904 (2016).
Hattne, J., Shi, D., de la Cruz, M. J., Reyes, F. E. & Gonen, T. Modeling truncated pixel values of faint reflections in MicroED images. J. Appl. Crystallogr. 49, 1029–1034 (2016).
Kabsch, W. XDS. Acta Crystallogr. D. Biol. Crystallogr. 66, 125–132 (2010).
Sheldrck, G. M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Delano, W.L. The PyMOL Molecular Graphics System (DeLano Scientific, 2002).
McGaughey, G. B., Gagné, M. & Rappé, A. K. π-Stacking interactions. Alive and well in proteins. J. Biol. Chem. 273, 15458–15463 (1998).
McDonald, I. K. & Thornton, J. M.Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 238, 777–793 (1994).
Acknowledgements
We thank C. Ophus (Molecular Foundry-NCEM), J. Miao (UCLA), and H. Nelson (UCLA) for helpful discussions. This work is supported by DOE Grant DE-FC02-02ER63421, the EICN in the CNSI at UCLA, the Janelia Research Visitor Program, and the NE-CAT beamline 24-ID-E, funded by NIH-NIGMS P41 GM103403. This work was also supported by STROBE: a National Science Foundation Science and Technology Center under Grant No. DMR 1548924. G.F.H. is supported by the BEC.AR program, Fundación Bunge y Born, Fundación Williams, and Fundación René Barón, and is a member of CONICET. M.G.J. is supported by a QCB Collaboratory Postdoctoral Fellowship. C.G. is supported by the Cellular and Molecular Biology Training Program. J.A.R. is supported by the Searle Scholar Program and the Beckman Young Investigator Program. T.G. and D.S.E. are supported by the Howard Hughes Medical Institute (HHMI), and J.E.E. and I.V.N. are supported by the DOE-BER Molecules to Mesoscale Bioimaging Project FWP #66382.
Author information
Authors and Affiliations
Contributions
J.A.R. directed the work. J.A.R., J.M., E.H., M.W.M., and C.G. grew, evaluated, and optimized crystals. J.A.R., D.R.B., C.G., M.R.S., H.T.M., M.G.-J., C.-T.Z., I.V.N., J.E.E., and D.C. collected data. J.A.R., C.G., J.M., M.G.-J., M.R.S., D.C., M.W.M., G.F.H., E.H., L.G., D.S.E., and T.G. analyzed the data. C.G., M.G.-J., and J.A.R. wrote the article, with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Sequence analysis of the common core of prionprotofilaments.
a, Tightly packed prion core as defined by NMR (Zahn, R. et al. NMR solution structure of the human prion protein. Proc. Natl. Acad. Sci. 97, 145–150 (2000)), Proteinase K resistance (Bocharova, O. V., Breydo, L., Salnikov, V. V., Gill, A. C. & Baskakov, I. V. Synthetic prions generated in vitro are similar to a newly identified subpopulation of PrPsc from sporadic Creutzfeldt-Jakob Disease. Protein Sci. Publ. Protein Soc. 14, 1222–1232 (2005); Bocharova, O. V. et al. Annealing Prion Protein Amyloid Fibrils at High Temperature Results in Extension of a Proteinase K-resistant Core. J. Biol. Chem. 281, 2373–2379 (2006)), Hydrogen-Deuterium Exchange (Dutta, A., Chen, S. & Surewicz, W. K. The effect of β2-α2 loop mutation on amyloidogenic properties of the prion protein. FEBS Lett. 587, 2918 (2013); Smirnovas, V. et al. Distinct Structures of Scrapie Prion Protein (PrPSc)-seeded Versus Spontaneous Recombinant Prion Protein Fibrils Revealed by Hydrogen/Deuterium Exchange. J. Biol. Chem. 284, 24233–24241 (2009); Lu, X., Wintrode, P. L. & Surewicz, W. K. β-Sheet core of human prion protein amyloid fibrils as determined by hydrogen/deuterium exchange. Proc. Natl. Acad. Sci. 104, 1510–1515 (2007); Cobb, N. J., Apostol, M. I., Chen, S., Smirnovas, V. & Surewicz, W. K. Conformational Stability of Mammalian Prion Protein Amyloid Fibrils Is Dictated by a Packing Polymorphism within the Core Region. J. Biol. Chem. 289, 2643–2650 (2014).), and EPR (Cobb, N. J., Sönnichsen, F. D., Mchaourab, H. & Surewicz, W. K. Molecular architecture of human prion protein amyloid: A parallel, in-register β-structure. Proc. Natl. Acad. Sci. U. S. A. 104, 18946–18951 (2007)). b, sequence alignment of bank vole PrP 27-30 with the 19 most similar sequences containing the proto-PrPScsequence (top, blue) and commonly studied PrP sequences (bottom, green) where only deviations from the bank vole sequence are shown.
Supplementary Figure 2 Characterization of proto-PrPSc aggregates.
a, MALDI-TOF/TOF mass spectrum of proto-PrPSc (Exact Mass of 1140.5 Da). b, Detection of Proto-PrPSc aggregates at an optical wavelength of 350 nm. Colors indicate solutions with increasing peptide concentrations with optical density plotted over the course of several hours. Values represent the mean OD 350 values and error bars represent ± 1 standard deviation (n = 3, technical replicates). c, Light microscope images of peptide solutions in a 96-well plate and electron micrographs of samples from those wells measured after 3 and 6 hours of incubation. d, Fiber diffraction with 10.2Å, 4.9Å, and 2.5Å rings labeled. Inset shows optical microscope image of the fibril from which diffraction was obtained.
Supplementary Figure 3 Physical and chemical denaturation of proto-PrPSc aggregates.
Visible light spectra (250-700nm) show absorbance from proto-PrPSc aggregates exposed to: a, a range of pH b, concentrations of guanidinium-HCl and c, concentrations of urea. Insets show each sample in a glass capillary above its corresponding figure legend. A 0.01% (w/v) solution of one µm latex spheres (grey lines) was used as a baseline against which to gauge particle density in various samples. The mean value of three replicates is shown (solid lines) with the shadowed range around each line representing ± 1 standard deviation (n = 3, technical replicates).
Supplementary Figure 4 Microfocal x-ray structure determination of proto-PrPSc and comparison to MicroED structure of proto-PrPSc.
a, Diffraction image collected by microfocal x-ray diffraction. Lower right inset shows hanging drop with a cluster of microcrystals (white arrow). Blue box in upper right shows a magnified view of high-resolution reflections near 1.1Å (black arrows) from the lower quadrant. b, Structure of proto-PrPSc determined by microfocal x-ray crystallography shown as a stick model. Waters are shown as cross marks, 2Fo-Fc density is shown at 1σ (blue mesh) and Fo-Fc at 2.8σ (green isosurface). c, Alignment of microfocal x-ray (blue) and MicroED (purple) structures. The peptide backbone is represented as an idealized beta strand and side chains as stick models. Residues are labeled according to their numbering in the sequence of bank vole prion.
Supplementary Figure 5 Ab initio structure determination of proto-PrPSc.
CFOM distribution obtained after 50,000 idenpendent trials of the SHELXD program. The ab initio model (top inset) results from the best CFOM in this distribution; the final solution is shown below. The blue box (CFOM > 80) defines regions where correct solutions are expected. The lower insets demonstrate the need to modify hydrogen positions from their idealized placement (magenta arrow) to better match the residual density (green arrow). Hydrogen bonding positions for the final model are shown as dashed lines.
Supplementary Figure 6 B factors in proto-PrPSc.
a, Side and b, top view of an isosurface model of proto-PrPSc analogous to those in Figure 1, but now colored by B-factor, where blue indicates low B-factors while red indicates high B-factors. A stick model is shown overlaid onto the isosurface model as a guide. The B-factor scale ranges from 0 to 18.0Å2.
Supplementary Figure 7 Hydrogen bond networks formed by polar side chains in β-sheets.
Five examples of hydrogen bonding networks are shown. From left to right, they are the proton wire (Nagle, J. F. & Morowitz, H. J. Molecular mechanisms for proton transport in membranes. Proc. Natl. Acad. Sci. U. S. A. 75, 298–302 (1978).), polar zipper (Perutz, M. F., Staden, R., Moens, L. & De Baere, I. Polar zippers. Curr. Biol. CB 3, 249–253 (1993).), glutamine ladder (Perutz, M. F., Johnson, T., Suzuki, M. & Finch, J. T. Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc. Natl. Acad. Sci. U. S. A. 91, 5355–5358 (1994).), asparagine ladder (Yoder, M. D., Lietzke, S. E. & Jurnak, F. Unusual structural features in the parallel β-helix in pectate lyases. Structure 1, 241–251 (1993).), and the polar clasp (blue outline). For each, three layers are shown, each representing three beta strands stacked along a sheet. Hydrogen bonds are shown as dashed lines.
Supplementary Figure 8 Refinement of proto-PrPSc as a function of resolution.
High resolution refinement of proto-PrPSc allows visualization of density for hydrogen atoms in density maps (orange arrows) at 0.7σ; the same atoms cannot be seen at lower resolution. Bridging density for the hydrogen bond between Q172 and N174 (blue arrows) can similarly only be seen at sub-ångstrom resolution.
Supplementary Figure 9 Comparison of ultrahigh-resolution features in maps solved by MicroED maps and X-ray crystallography.
Comparison of ultrahigh resolution features in MicroED maps of proto-PrPSc to those of 0.48Å and 0.8Å x-ray crystallographic maps of the hiPiP protein (PDB IDs: 1IUA and 5D8V) (Hirano, Y., Takeda, K. & Miki, K. Charge-density analysis of an iron sulfur protein at an ultra-high resolution of 0.48 Å. Nature 534, 281–284 (2016); Liu, L., Nogi, T., Kobayashi, M., Nozawa, T. & Miki, K. Ultrahigh-resolution structure of high-potential iron sulfur protein from Thermochromatium tepidum. Acta Crystallogr. D Biol. Crystallogr. 58, 1085–1091 (2002).). 2mFoDFc maps are rendered as a blue mesh (0.8σ) and mFo-DFc maps are rendered as a solid green isosurface (2.8σ). Density for hydrogen atoms are indicated by red arrows and residual density in the difference map is indicated by blue arrows. The sidechain panel also includes contour maps for 2Fo-Fc and Fo-Fc density.
Supplementary Figure 10 Ab initio structure determination of carbamazepine.
CFOM distribution obtained after 100,000 independent trials of the SHELXD program. The ab initio model (top inset) results from the best CFOM in this distribution; the final solution is shown below. 2Fo-Fc maps are shown in blue, Fo-Fc maps in green inform the locations of hydrogens in the model. The blue box (CFOM > 80) defines the region where correct solutions are expected.
Supplementary Figure 11 Comparison of ultrahigh-resolution features in MicroED maps of proto-PrPSc refined in different software packages.
MicroED maps of proto-PrPSc were refined by either the Phenix (a and c) or Refmac (b and d) software packages. 2mFo-DFc maps are rendered as a blue mesh (0.8σ) and mFo-DFc maps are rendered as a solid green isosurface (2.8σ). Arrows indicate bridging hydrogen bonds. b, Fourier shell correlation between the maps produced by each refinement package, with a dashed black line indicating FSC cutoffs of 0.5 and 0.143.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11 and Supplementary Tables 1–4
Rights and permissions
About this article
Cite this article
Gallagher-Jones, M., Glynn, C., Boyer, D.R. et al. Sub-ångström cryo-EM structure of a prion protofibril reveals a polar clasp. Nat Struct Mol Biol 25, 131–134 (2018). https://doi.org/10.1038/s41594-017-0018-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-017-0018-0
This article is cited by
-
Hampering the early aggregation of PrP-E200K protein by charge-based inhibitors: a computational study
Journal of Computer-Aided Molecular Design (2021)
-
MyD88 TIR domain higher-order assembly interactions revealed by microcrystal electron diffraction and serial femtosecond crystallography
Nature Communications (2021)
-
Cryo-EM structure of a human prion fibril with a hydrophobic, protease-resistant core
Nature Structural & Molecular Biology (2020)
-
Cryo-EM structure of an amyloid fibril formed by full-length human prion protein
Nature Structural & Molecular Biology (2020)
-
Nanoscale mosaicity revealed in peptide microcrystals by scanning electron nanodiffraction
Communications Biology (2019)