Abstract
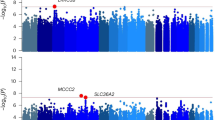
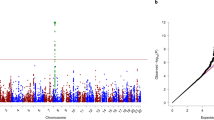
Cannabis use is a heritable trait that has been associated with adverse mental health outcomes. In the largest genome-wide association study (GWAS) for lifetime cannabis use to date (N = 184,765), we identified eight genome-wide significant independent single nucleotide polymorphisms in six regions. All measured genetic variants combined explained 11% of the variance. Gene-based tests revealed 35 significant genes in 16 regions, and S-PrediXcan analyses showed that 21 genes had different expression levels for cannabis users versus nonusers. The strongest finding across the different analyses was CADM2, which has been associated with substance use and risk-taking. Significant genetic correlations were found with 14 of 25 tested substance use and mental health–related traits, including smoking, alcohol use, schizophrenia and risk-taking. Mendelian randomization analysis showed evidence for a causal positive influence of schizophrenia risk on cannabis use. Overall, our study provides new insights into the etiology of cannabis use and its relation with mental health.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
05 June 2019
Several occurrences of the word ‘schizophrenia’ have been re-worded as ‘liability to schizophrenia’ or ‘schizophrenia risk’, including in the title, which should have been “GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal effect of schizophrenia liability,” as well as in Supplementary Figures 1–10 and Supplementary Tables 7–10, to more accurately reflect the findings of the work.
References
Volkow, N. D., Compton, W. M. & Weiss, S. R. Adverse health effects of marijuana use. N. Engl. J. Med. 371, 879 (2014).
Moore, T. H. et al. Cannabis use and risk of psychotic or affective mental health outcomes: a systematic review. Lancet 370, 319–328 (2007).
Hall, W. & Degenhardt, L. Adverse health effects of non-medical cannabis use. Lancet 374, 1383–1391 (2009).
Verweij, K. J. H. et al. Genetic and environmental influences on cannabis use initiation and problematic use: a meta-analysis of twin studies. Addiction 105, 417–430 (2010).
Agrawal, A., Neale, M. C., Jacobson, K. C., Prescott, C. A. & Kendler, K. S. Illicit drug use and abuse/dependence: modeling of two-stage variables using the CCC approach. Addict. Behav. 30, 1043–1048 (2005).
Agrawal, A. & Lynskey, M. T. The genetic epidemiology of cannabis use, abuse and dependence. Addiction 101, 801–812 (2006).
Verweij, K. J. H. et al. The genetic aetiology of cannabis use initiation: a meta-analysis of genome-wide association studies and a SNP-based heritability estimation. Addict. Biol. 18, 846–850 (2013).
Agrawal, A. et al. A genome-wide association study of DSM-IV cannabis dependence. Addict. Biol. 16, 514–518 (2011).
Minica, C. C. et al. Heritability, SNP- and gene-based analyses of cannabis use initiation and age at onset. Behav. Genet. 45, 503–513 (2015).
Stringer, S. et al. Genome-wide association study of lifetime cannabis use based on a large meta-analytic sample of 32 330 subjects from the International Cannabis Consortium. Transl. Psychiatry 6, e769 (2016).
Demontis, D. et al. Genome-wide association study implicates CHRNA2 in cannabis use disorder. Preprint at bioRxiv https://doi.org/10.1101/237321 (2018).
Gelernter, J. et al. Haplotype spanning TTC12 and ANKK1, flanked by the DRD2 and NCAM1 loci, is strongly associated to nicotine dependence in two distinct American populations. Hum. Mol. Genet. 15, 3498–3507 (2006).
Yang, B. Z. et al. Association of haplotypic variants in DRD2, ANKK1, TTC12 and NCAM1 to alcohol dependence in independent case control and family samples. Hum. Mol. Genet. 16, 2844–2853 (2007).
Clarke, T.-K. et al. Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N = 112 117). Mol. Psychiatry 22, 1376–1384 (2017).
Boutwell, B. et al. Replication and characterization of CADM2 and MSRA genes on human behavior. Heliyon 3, e00349 (2017).
Day, F. R. et al. Physical and neurobehavioral determinants of reproductive onset and success. Nat. Genet. 48, 617–623 (2016).
Andréasson, S., Allebeck, P., Engström, A. & Rydberg, U. Cannabis and schizophrenia. A longitudinal study of Swedish conscripts. Lancet 2, 1483–1486 (1987).
Smit, F., Bolier, L. & Cuijpers, P. Cannabis use and the risk of later schizophrenia: a review. Addiction 99, 425–430 (2004).
Volkow, N. D. et al. Effects of cannabis use on human behavior, including cognition, motivation, and psychosis: a review. JAMA Psychiatry 73, 292–297 (2016).
Verweij, K. J. et al. Short communication: Genetic association between schizophrenia and cannabis use. Drug Alcohol Depend. 171, 117–121 (2017).
Power, R. A. et al. Genetic predisposition to schizophrenia associated with increased use of cannabis. Mol. Psychiatry 19, 1201–1204 (2014).
Burgess, S., Scott, R. A., Timpson, N. J., Davey Smith, G. & Thompson, S. G. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur. J. Epidemiol. 30, 543–552 (2015).
Vaucher, J. et al. Cannabis use and risk of schizophrenia: a Mendelian randomization study. Mol. Psychiatry 23, 1287–1292 (2018).
Gage, S. H. et al. Assessing causality in associations between cannabis use and schizophrenia risk: a two-sample Mendelian randomization study. Psychol. Med. 47, 971–980 (2017).
de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLOS Comput. Biol. 11, e1004219 (2015).
Barbeira, A.N. et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Preprint at bioRxiv https://doi.org/10.1101/045260 (2017).
Bowden, J., Davey Smith, G. & Burgess, S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525 (2015).
Burgess, S. & Thompson, S. G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 32, 377–389 (2017).
Ibrahim-Verbaas, C. A. et al. GWAS for executive function and processing speed suggests involvement of the CADM2 gene. Mol. Psychiatry 21, 189–197 (2016).
Walther, B., Morgenstern, M. & Hanewinkel, R. Co-occurrence of addictive behaviours: personality factors related to substance use, gambling and computer gaming. Eur. Addict. Res. 18, 167–174 (2012).
Martin, C. A. et al. Sensation seeking, puberty, and nicotine, alcohol, and marijuana use in adolescence. J. Am. Acad. Child Adolesc. Psychiatry 41, 1495–1502 (2002).
Nielsen, J., Kulahin, N. & Walmod, P. S. Extracellular protein interactions mediated by the neural cell adhesion molecule, NCAM: heterophilic interactions between NCAM and cell adhesion molecules, extracellular matrix proteins, and viruses. Adv. Exp. Med. Biol. 663, 23–53 (2010).
Rubinek, T. et al. The cell adhesion molecules N-cadherin and neural cell adhesion molecule regulate human growth hormone: a novel mechanism for regulating pituitary hormone secretion. J. Clin. Endocrinol. Metab. 88, 3724–3730 (2003).
Ducci, F. et al. TTC12-ANKK1-DRD2 and CHRNA5-CHRNA3-CHRNB4 influence different pathways leading to smoking behavior from adolescence to mid-adulthood. Biol. Psychiatry 69, 650–660 (2011).
Bidwell, L. C. et al. NCAM1-TTC12-ANKK1-DRD2 variants and smoking motives as intermediate phenotypes for nicotine dependence. Psychopharmacology (Berl.) 232, 1177–1186 (2015).
Atz, M. E., Rollins, B. & Vawter, M. P. NCAM1 association study of bipolar disorder and schizophrenia: polymorphisms and alternatively spliced isoforms lead to similarities and differences. Psychiatr. Genet. 17, 55–67 (2007).
Petrovska, J. et al. The NCAM1 gene set is linked to depressive symptoms and their brain structural correlates in healthy individuals. J. Psychiatr. Res. 91, 116–123 (2017).
Weiss, L. A. et al. Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 358, 667–675 (2008).
McCarthy, S. E. et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat. Genet. 41, 1223–1227 (2009).
González, J. R. et al. A common 16p11.2 inversion underlies the joint susceptibility to asthma and obesity. Am. J. Hum. Genet. 94, 361–372 (2014).
Zuo, L. et al. Genome-wide significant association signals in IPO11-HTR1A region specific for alcohol and nicotine codependence. Alcohol. Clin. Exp. Res. 37, 730–739 (2013).
Speliotes, E. K. et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 42, 937–948 (2010).
Donaldson, Z. R. et al. The functional serotonin 1a receptor promoter polymorphism, rs6295, is associated with psychiatric illness and differences in transcription. Transl. Psychiatry 6, e746 (2016).
Gatt, J. M., Burton, K. L., Williams, L. M. & Schofield, P. R. Specific and common genes implicated across major mental disorders: a review of meta-analysis studies. J. Psychiatr. Res. 60, 1–13 (2015).
Takekita, Y. et al. HTR1A polymorphisms and clinical efficacy of antipsychotic drug treatment in schizophrenia: a meta-analysis. Int. J. Neuropsychopharmacol. 19, pyv125 (2016).
Sherva, R. et al. Genome-wide association study of cannabis dependence severity, novel risk variants, and shared genetic risks. JAMA Psychiatry 73, 472–480 (2016).
Patrick, M. E., Wightman, P., Schoeni, R. F. & Schulenberg, J. E. Socioeconomic status and substance use among young adults: a comparison across constructs and drugs. J. Stud. Alcohol Drugs 73, 772–782 (2012).
Müller-Vahl, K. R. & Emrich, H. M. Cannabis and schizophrenia: towards a cannabinoid hypothesis of schizophrenia. Expert Rev. Neurother. 8, 1037–1048 (2008).
Abecasis, G. R. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012).
McCarthy, S. & Das, S. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 48, 1279–1283 (2016).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Winkler, T. W. & Day, F. R. Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 9, 1192–1212 (2014).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Turner, S.D. qqman: an R package for visualizing GWAS results using Q-Q and Manhattan plots. Preprint at bioRxiv https://doi.org/10.1101/005165 (2014).
Pruim, R. J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010).
Gamazon, E. R. & Wheeler, H. E. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 47, 1091–1098 (2015).
Delaneau, O. & Marchini, J. Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat. Commun. 5, 3934 (2014).
Carithers, L. J. et al. A novel approach to high-quality postmortem tissue procurement: the GTEx project. Biopreserv. Biobank. 13, 311–319 (2015).
Sul, J. H., Han, B., Ye, C., Choi, T. & Eskin, E. Effectively identifying eQTLs from multiple tissues by combining mixed model and meta-analytic approaches. PLoS Genet. 9, e1003491 (2013).
Bulik-Sullivan, B. K. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Altshuler, D. M. et al. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Hemani, G. et al. MR-Base: a platform for systematic causal inference across the phenome using billions of genetic associations. Preprint at bioRxiv https://doi.org/10.1101/078972 (2017).
Zhu, Z. et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat. Commun. 9, 224 (2018).
Davey Smith, G. & Hemani, G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 23(R1), R89–R98 (2014).
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Ehret, G. B. et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 478, 103–109 (2011).
Bowden, J., Davey Smith, G., Haycock, P. C. & Burgess, S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40, 304–314 (2016).
Hartwig, F.P., Davey Smith, G. & Bowden, J. Robust inference in two-sample Mendelian randomisation via the zero modal pleiotropy assumption. Preprint at bioRxiv https://doi.org/10.1101/126102 (2017).
Bowden, J. et al. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I 2 statistic. Int. J. Epidemiol. 45, 1961–1974 (2016).
Acknowledgements
We would like to thank the research participants and employees of 23andMe for making this work possible. We gratefully acknowledge the Psychiatric Genomics Consortium contributing studies and the participants in those studies without whom this effort would not have been possible. J.A.P. and J.M.V. are supported by the European Research Council (Beyond the Genetics of Addiction ERC-284167, PI J.M.V.). K.J.H.V. is supported by the Foundation Volksbond Rotterdam. N.A.G. is supported by US National Institutes of Health, National Institute on Drug Abuse R00DA023549. J.L.T. is supported by the Netherlands Organization for Scientific Research (NWO; Rubicon grant 446-16-009). S.M. is supported by an Australian Research Council Fellowship. Statistical analyses were partly carried out on the Genetic Cluster Computer (http://www.geneticcluster.org) hosted by SURFsara and financially supported by the Netherlands Organization for Scientific Research (NWO 480-05-003 PI: Posthuma) along with a supplement from the Dutch Brain Foundation and the VU University Amsterdam. M.G.N. is supported by Royal Netherlands Academy of Science Professor Award to D.I.B. (PAH/6635). Part of the computation of this project was funded by NWO Exact Sciences for the application: “Population scale Genetic Analysis” awarded to M.G.N. The genome-wide association analysis on the UK Biobank dataset has been conducted using the UK Biobank resource under application numbers 9905, 16406 and 25331.
The Substance Use Disorders Working Group of the Psychiatric Genomics Consortium (PGC-SUD) is supported by funds from NIDA and NIMH to MH109532 and, previously, with analyst support from NIAAA to U01AA008401 (COGA). J.M.’s contributions were partially supported by the Peter Boris Chair in Addictions Research. S.S.-R. was supported by the Frontiers of Innovation Scholars Program (FISP; #3-P3029), the Interdisciplinary Research Fellowship in NeuroAIDS (IRFN; MH081482) and a pilot award from DA037844. R.M. was supported by the European Union through the European Regional Development Fund (Project No. 2014-2020.4.01.15-0012) and the European Union’s Horizon 2020 Research and Innovation Programme under grant agreements No 692065 and 692145. J.K. was supported by Academy Professorship grants by the Academy of Finland (263278, 292782). M.R. is a recipient of a Miguel de Servet contract from the Instituto de Salud Carlos III, Spain (CP09/00119 and CPII15/00023).
Contributors to the 23andMe Research Team
M. Agee, B. Alipanahi, A. Auton, R. K. Bell, K. Bryc, S. L. Elson, P. Fontanillas, N. A. Furlotte, D. A. Hinds, K. E. Huber, A. Kleinman, N. K. Litterman, J. C. McCreight, M. H. McIntyre, J. L. Mountain, E. S. Noblin, C. A. M. Northover, S. J. Pitts, J. Fah Sathirapongsasuti, O. V. Sazonova, J. F. Shelton, S. Shringarpure, C. Tian, J. Y. Tung, V. Vacic and C.H. Wilson.
Contributors to the International Cannabis Consortium
S. Stringer, C. C. Minica, K. J. H. Verweij, H. Mbarek, M. Bernard, J. Derringer, K. R. van Eijk, J. D. Isen, A. Loukola, D. F. Maciejewski, E. Mihailov, P. J. van der Most, C. Sánchez-Mora, L. Roos, R. Sherva, R. Walters, J.J. Ware, A. Abdellaoui, T. B. Bigdeli, S. J. T. Branje, S. A. Brown, M. Bruinenberg, M. Casas, T. Esko, I. Garcia-Martinez, S. D. Gordon, J. M. Harris, C. A. Hartman, A. K. Henders, A. C. Heath, I. B. Hickie, M. Hickman, C. J. Hopfer, J. J. Hottenga, A. C. Huizink, D. E. Irons, R. S. Kahn, T. Korhonen, H. R. Kranzler, K. Krauter, P. A. C. van Lier, G. H. Lubke, P. A. F. Madden, R. Mägi, M. K. McGue, S. E. Medland, W. H. J. Meeus, M. B. Miller, G. W. Montgomery, M. G. Nivard, I. M. Nolte, A. J. Oldehinkel, Z. Pausova, B. Qaiser, L. Quaye, J. A. Ramos-Quiroga, V. Richarte, R. J. Rose, J. Shin, M. C. Stallings, A. I. Stiby, T. L. Wall, M. J. Wright, H. M. Koot, T. Paus, J. K. Hewitt, M. Ribasés, J. Kaprio, M. P. M. Boks, H. Snieder, T. Spector, M. R. Munafò, A. Metspalu, J. Gelernter, D. I. Boomsma, W. G. Iacono, N. G. Martin, N. A. Gillespie, E. M. Derks and J. M. Vink.
Author information
Authors and Affiliations
Consortia
Contributions
K.J.H.V., E.M.D. and J.M.V. were responsible for the study concept and the design of the study. I.C.C. contributed existing genome-wide summary data from the International Cannabis Consortium. The PGC-SUD group provided summary statistics of the cannabis dependence GWAS. S.S., S.S.-R., M.G.N., B.M.L.B., J.-S.O., H.F.I., M.D.v.d.Z., M.B., F.R.D., P.F., S.M., J.R.B.P., A.A.P. and D.P. performed or supervised genome-wide association analyses. J.A.P. performed the quality control and meta-analysis of genome-wide association studies, under supervision of K.J.H.V., B.M.L.B. and J.M.V. J.A.P., K.J.H.V., Z.G., J.L.T., A.A., M.R.M. and E.M.D. contributed to secondary analyses of the data. J.A.P., K.J.H.V., Z.G., N.A.G., E.M.D. and J.M.V. wrote the manuscript. J.L.D., S.J.T.B., C.A.H., A.C.H., P.A.C.v.L., P.A.F.M., R.M., W.M., G.W.M., A.J.O., Z.P., J.A.R.-Q., T.P., M.R., J.K., M.P.M.B., J.T.B., T.D.S., J.G., D.I.B. and N.G.M. contributed to data acquisition of the samples in the International Cannabis Consortium. S.L.E., H.d.W., L.K.D. and J.M.K. contributed to data acquisition and analysis for the 23andMe dataset. All authors provided critical revision of the manuscript for important intellectual content.
Corresponding author
Ethics declarations
Competing interests
P.F., S.L.E. and members of the 23andMe Research Team are employees of 23andMe Inc. J.A.R.-Q. was on the speakers’ bureau and/or acted as consultant for Eli Lilly, Janssen-Cilag, Novartis, Shire, Lundbeck, Almirall, BRAINGAZE, Sincrolab and Rubió in the last 5 years. He also received travel awards (air tickets and hotel) for taking part in psychiatric meetings from Janssen-Cilag, Rubió, Shire and Eli Lilly. The Department of Psychiatry chaired by him received unrestricted educational and research support from the following pharmaceutical companies in the last 5 years: Eli Lilly, Lundbeck, Janssen- Cilag, Actelion, Shire, Ferrer and Rubió.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10
Supplementary Table 1
All genome-wide significant SNP associations in the meta-analysis
Supplementary Table 2
Independent genome-wide significant associations with lifetime cannabis use in the UK Biobank sample
Supplementary Table 3
Description of the genome-wide significant associations in the gene-based test of association and the S-PrediXcan analysis, with a short (non-comprehensive) overview of relevant literature findings on gene–phenotype associations
Supplementary Table 4
Significant S-PrediXcan associations after correction for multiple testing
Supplementary Table 5
Summary of S-PrediXcan associations by target gene
Supplementary Table 6
Results from LD score regression analysis: genetic correlations between lifetime cannabis use and various traits of interest
Supplementary Table 7
SNPs included in the genetic instruments used for bidirectional two-sample Mendelian randomization analyses between lifetime cannabis use and schizophrenia diagnosis
Supplementary Table 8
Cochran’s heterogeneity statistic (Q) for inverse-variance-weighted (IVW) bidirectional two-sample Mendelian randomization analyses between lifetime cannabis use and schizophrenia diagnosis
Supplementary Table 9
I2 statistic for the heterogeneity between genetic variants in an instrument for the MR-Egger SIMEX analysis
Supplementary Table 10
MR-Egger SIMEX intercept, indicating degree of horizontal pleiotropy, for bidirectional two-sample Mendelian randomization analyses between lifetime cannabis use and schizophrenia diagnosis
Supplementary Table 11
Methodological details of the individual GWASs
Supplementary Table 12
quality control steps in the individual GWASs
Rights and permissions
About this article
Cite this article
Pasman, J.A., Verweij, K.J.H., Gerring, Z. et al. GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal effect of schizophrenia liability. Nat Neurosci 21, 1161–1170 (2018). https://doi.org/10.1038/s41593-018-0206-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-018-0206-1
This article is cited by
-
Development and initial validation of the cannabis-related psychosis risk literacy scale (CPRL): a multinational psychometric study
BMC Psychiatry (2024)
-
Epidemiologic and genetic associations of female reproductive disorders with depression or dysthymia: a Mendelian randomization study
Scientific Reports (2024)
-
Genetic influences and causal pathways shared between cannabis use disorder and other substance use traits
Molecular Psychiatry (2024)
-
Pleiotropy and genetically inferred causality linking multisite chronic pain to substance use disorders
Molecular Psychiatry (2024)
-
The causal impact of mental health on tobacco and alcohol consumption: an instrumental variables approach
Empirical Economics (2024)