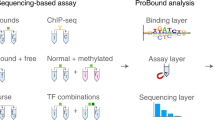
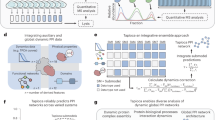
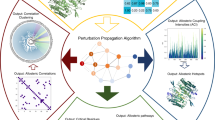
Abstract
In mammalian cells, much of signal transduction is mediated by weak protein–protein interactions between globular peptide-binding domains (PBDs) and unstructured peptidic motifs in partner proteins. The number and diversity of these PBDs (over 1,800 are known), their low binding affinities and the sensitivity of binding properties to minor sequence variation represent a substantial challenge to experimental and computational analysis of PBD specificity and the networks PBDs create. Here, we introduce a bespoke machine-learning approach, hierarchical statistical mechanical modeling (HSM), capable of accurately predicting the affinities of PBD–peptide interactions across multiple protein families. By synthesizing biophysical priors within a modern machine-learning framework, HSM outperforms existing computational methods and high-throughput experimental assays. HSM models are interpretable in familiar biophysical terms at three spatial scales: the energetics of protein–peptide binding, the multidentate organization of protein–protein interactions and the global architecture of signaling networks.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The domain–peptide and PPI predictions are made available through a custom website (https://ProteinPeptide.io). The protein–peptide interaction data are also made available in figshare with the identifiers https://doi.org/10.6084/m9.figshare.10084745. Data used in training the model are available as Supplementary Dataset 2.
Code availability
All code and data used for training and testing HSM are available in a public repository at https://github.com/aqlaboratory/hsm.
References
Gao, A. et al. Evolution of weak cooperative interactions for biological specificity. Proc. Natl Acad. Sci. USA 115, E11053–E11060 (2018).
Perkins, J. R., Diboun, I., Dessailly, B. H., Lees, J. G. & Orengo, C. Transient protein–protein interactions: structural, functional, and network properties. Structure 18, 1233–1243 (2010).
Mayer, B. J. The discovery of modular binding domains: building blocks of cell signalling. Nat. Rev. Mol. Cell Biol. 16, 691–698 (2015).
Tompa, P., Davey, N. E., Gibson, T. J. & Babu, M. M. A million peptide motifs for the molecular biologist. Mol. Cell 55, 161–169 (2014).
Scott, J. D. & Pawson, T. Cell signaling in space and time: where proteins come together and when they’re apart. Science 326, 1220–1224 (2009).
Cesareni, G., Gimona, M., Sudol, M. & Yaffe, M. Modular Protein Domains (John Wiley & Sons, 2006).
Yang, F. et al. Protein domain-level landscape of cancer-type-specific somatic mutations. PLoS Comput. Biol. 11, e1004147 (2015).
Miller, M. L. et al. Pan-cancer analysis of mutation hotspots in protein domains. Cell Syst. 1, 197–209 (2015).
Rual, J.-F. et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature 437, 1173–1178 (2005).
Beck, M., Claassen, M. & Aebersold, R. Comprehensive proteomics. Curr. Opin. Biotechnol. 22, 3–8 (2011).
Hein, M. Y. et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163, 712–723 (2015).
Levinson, N. M., Seeliger, M. A., Cole, P. A. & Kuriyan, J. Structural basis for the recognition of c-Src by its inactivator Csk. Cell 134, 124–134 (2008).
Waksman, G., Shoelson, S. E., Pant, N., Cowburn, D. & Kuriyan, J. Binding of a high affinity phosphotyrosyl peptide to the Src SH2 domain: crystal structures of the complexed and peptide-free forms. Cell 72, 779–790 (1993).
Demers, J.-P. & Mittermaier, A. Binding mechanism of an SH3 domain studied by NMR and ITC. J. Am. Chem. Soc. 131, 4355–4367 (2009).
Tinti, M. et al. The SH2 domain interaction landscape. Cell Rep. 3, 1293–1305 (2013).
Hou, T., Chen, K., McLaughlin, W. A., Lu, B. & Wang, W. Computational analysis and prediction of the binding motif and protein interacting partners of the Abl SH3 domain. PLoS Comput. Biol. 2, e1 (2006).
Kundu, K., Mann, M., Costa, F. & Backofen, R. MoDPepInt: an interactive web server for prediction of modular domain–peptide interactions. Bioinformatics 30, 2668–2669 (2014).
Mignon, D., Panel, N., Chen, X., Fuentes, E. J. & Simonson, T. Computational design of the Tiam1 PDZ domain and its ligand binding. J. Chem. Theory Comput. 13, 2271–2289 (2017).
Kaneko, T. et al. Loops govern SH2 domain specificity by controlling access to binding pockets. Sci. Signal 3, ra34 (2010).
AlQuraishi, M., Koytiger, G., Jenney, A., MacBeath, G. & Sorger, P. K. A multiscale statistical mechanical framework integrates biophysical and genomic data to assemble cancer networks. Nat. Genet. 46, 1363–1372 (2014).
Schroeder, D. V. An Introduction to Thermal Physics (Addison-Wesley, 2000).
Goldstein, H., Poole Jr., C. P. & Safko, J. L. Classical Mechanics (Addison-Wesley, 2001).
AlQuraishi, M. & McAdams, H. H. Direct inference of protein–DNA interactions using compressed sensing methods. Proc. Natl Acad. Sci. USA 108, 14819–14824 (2011).
Zarrinpar, A., Bhattacharyya, R. P. & Lim, W. A. The structure and function of proline recognition domains. Sci. STKE 2003, re8 (2003).
Denu, J. M. & Dixon, J. E. Protein tyrosine phosphatases: mechanisms of catalysis and regulation. Curr. Opin. Chem. Biol. 2, 633–641 (1998).
Wagner, M. J., Stacey, M. M., Liu, B. A. & Pawson, T. Molecular mechanisms of SH2- and PTB-domain-containing proteins in receptor tyrosine kinase signaling. Cold Spring Harb. Perspect. Biol. 5, a008987 (2013).
Harris, B. Z. & Lim, W. A. Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci. 114, 3219–3231 (2001).
Kolodny, R., Koehl, P., Guibas, L. & Levitt, M. Small libraries of protein fragments model native protein structures accurately. J. Mol. Biol. 323, 297–307 (2002).
Nepomnyachiy, S., Ben-Tal, N. & Kolodny, R. Global view of the protein universe. Proc. Natl Acad. Sci. USA 111, 11691–11696 (2014).
Stormo, G. D., Schneider, T. D., Gold, L. & Ehrenfeucht, A. Use of the ‘Perceptron’ algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res. 10, 2997–3011 (1982).
Miller, M. L. et al. Linear motif atlas for phosphorylation-dependent signaling. Sci. Signal 1, ra2 (2008).
Chatr-aryamontri, A. et al. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 45, D369–D379 (2017).
Orchard, S. et al. The MIntAct project—IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 42, D358–D363 (2014).
Huttlin, E. L. et al. The bioplex network: a systematic exploration of the human interactome. Cell 162, 425–440 (2015).
Huttlin, E. L. et al. Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509 (2017).
Rolland, T. et al. A proteome-scale map of the human interactome network. Cell 159, 1212–1226 (2014).
Yoo, J., Lee, T.-S., Choi, B., Shon, M. J. & Yoon, T.-Y. Observing extremely weak protein–protein interactions with conventional single-molecule fluorescence microscopy. J. Am. Chem. Soc. 138, 14238–14241 (2016).
Lee, C. H. et al. A single amino acid in the SH3 domain of Hck determines its high affinity and specificity in binding to HIV-1 Nef protein. EMBO J. 14, 5006–5015 (1995).
Fernandez-Ballester, G., Blanes-Mira, C. & Serrano, L. The tryptophan switch: changing ligand-binding specificity from type I to type II in SH3 domains. J. Mol. Biol. 335, 619–629 (2004).
Schmidt, H. et al. Solution structure of a Hck SH3 domain ligand complex reveals novel interaction modes. J. Mol. Biol. 365, 1517–1532 (2007).
Teyra, J. et al. Comprehensive analysis of the human SH3 domain family reveals a wide variety of non-canonical specificities. Structure 25, 1598–1610.e3 (2017).
Ma’ayan, A. et al. Formation of regulatory patterns during signal propagation in a mammalian cellular network. Science 309, 1078–1083 (2005).
Goodfellow, I, Bengio, Y. & Courville, A. Deep Learning. (MIT Press, 2016).
Bengio, Y. Deep learning of representations for unsupervised and transfer learning. in Proc. ICML Workshop on Unsupervised and Transfer Learning Vol. 27 (eds Guyon, I. et al.) 17–36 (PMLR, 2012).
Snell, J., Swersky, K. & Zemel, R. S. in Advances in Neural Information Processing Systems Vol. 30 (eds Guyon, I. et al.) 4077–4087 (Curran Associates, Inc., 2017).
AlQuraishi, M. End-to-end differentiable learning of protein structure. Cell Syst. 8, 292–301.e3 (2019).
Xu, J. Distance-based protein folding powered by deep learning. Proc. Natl Acad. Sci. USA 116, 16856–16865 (2019).
Senior, A. W. et al. Protein structure prediction using multiple deep neural networks in the 13th Critical Assessment of Protein Structure Prediction (CASP13). Proteins 87, 1141–1148 (2019).
Kryshtafovych, A., Schwede, T., Topf, M., Fidelis, K. & Moult, J. Critical assessment of methods of protein structure prediction (CASP)—Round XIII. Proteins Struct. Funct. Bioinforma. 87, 1011–1020 (2019).
Wilson, D. et al. SUPERFAMILY—sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res. 37, D380–D386 (2009).
Sokal, R. R. & Michener, C. D. A statistical method for evaluating relationships. Univ. Kans. Sci. Bull. 38, 1409–1448 (1958).
DeLong, E. R., DeLong, D. M. & Clarke-Pearson, D. L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44, 837–845 (1988).
Hornbeck, P. V. et al. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 (2015).
Peng, J. & Xu, J. Raptorx: exploiting structure information for protein alignment by statistical inference. Proteins Struct. Funct. Bioinforma. 79, 161–171 (2011).
Dinkel, H. et al. ELM—the database of eukaryotic linear motifs. Nucleic Acids Res. 40, D242–D251 (2012).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 57, 289–300 (1995).
Peixoto, T. P. The Graph-Tool Python Library https://doi.org/10.6084/m9.figshare.1164194.v14 (2017).
Acknowledgements
This work was funded by NIH grants (nos. U54-CA225088 and P50-GM107618) and by DARPA/DOD (grant no. W911NF-14-1-0397) to P.K.S.
Author information
Authors and Affiliations
Contributions
J.M.C., P.K.S. and M.A. conceived and designed the model, analysis and computational experiments. J.M.C. implemented the model and carried out the analysis and experiments. G.K. collected and processed binding and structural data and contributed to the analysis. All authors wrote and reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
P.K.S. is a member of the SAB or Board of Directors of Merrimack Pharmaceutical, Glencoe Software, Applied Biomath and RareCyte Inc. and has equity in these companies. P.K.S. declares that none of these relationships are directly or indirectly related to the content of this manuscript.
Additional information
Peer review information Rita Strack was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–7, Tables 1–5 and Notes 1 and 2.
Supplementary Dataset 1
Domain sequences and multiple sequence alignments used in both HSM/D and HSM/P.
Supplementary Dataset 2
Raw domain-peptide training data used for training HSM/D.
Supplementary Dataset 3
Potential peptidic sites used in predictions generated with HSM/P.
Supplementary Dataset 4
Assessment of HSM/D relative to other domain models. Contains source data for Fig. 2a and Supplementary Fig. 3.
Supplementary Dataset 5
PyMOL structural data associated with analysing HSM/D inferred energy profiles. Includes source data for Figs. 4 and 5 and Supplementary Figs. 5 and 6.
Supplementary Dataset 6
Assessment of HSM/P. Contains source data for Figs. 2b, 3 and 6 and Supplementary Fig. 7.
Source data
Source Data Fig. 2
Source Data
Rights and permissions
About this article
Cite this article
Cunningham, J.M., Koytiger, G., Sorger, P.K. et al. Biophysical prediction of protein–peptide interactions and signaling networks using machine learning. Nat Methods 17, 175–183 (2020). https://doi.org/10.1038/s41592-019-0687-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-019-0687-1
This article is cited by
-
Machine learning for antimicrobial peptide identification and design
Nature Reviews Bioengineering (2024)
-
An E. coli display method for characterization of peptide–sensor kinase interactions
Nature Chemical Biology (2023)
-
Protein–protein interaction and non-interaction predictions using gene sequence natural vector
Communications Biology (2022)
-
PepNN: a deep attention model for the identification of peptide binding sites
Communications Biology (2022)
-
Generation of functional oligopeptides that promote osteogenesis based on unsupervised deep learning of protein IDRs
Bone Research (2022)