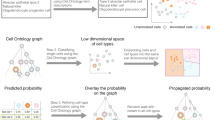
Abstract
Single-cell molecular profiling technologies are gaining rapid traction, but the manual process by which resulting cell types are typically annotated is labor intensive and rate-limiting. We describe Garnett, a tool for rapidly annotating cell types in single-cell transcriptional profiling and single-cell chromatin accessibility datasets, based on an interpretable, hierarchical markup language of cell type-specific genes. Garnett successfully classifies cell types in tissue and whole organism datasets, as well as across species.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
No new data was generated for this study. All data used in this study is publicly available.
References
Svensson, V., Vento-Tormo, R. & Teichmann, S. A. Nat. Protoc. 13, 599–604 (2018).
Tabula Muris Consortium Nature 562, 367–372 (2018).
Han, X. et al. Cell 173, 1307 (2018).
Zhang, X. et al. Nucleic Acids Res. 47, D721–D728 (2019).
Diehl, A. D. et al. J. Biomed. Semant. 7, 44 (2016).
Bard, J., Rhee, S. Y. & Ashburner, M. Genome Biol. 6, R21 (2005).
Cusanovich, D. A. et al. Cell 174, 1309–1324 (2018).
Zheng, G. X. Y. et al. Nat. Commun. 8, 14049 (2017).
Levine, J. H. et al. Cell 162, 184–197 (2015).
Pliner, H. A. et al. Mol. Cell 71, 858–871 (2018).
Cao, J. et al. Science 357, 661–667 (2017).
Zeisel, A. et al. Cell 174, 999–1014.e22 (2018).
Merkin, J., Russell, C., Chen, P. & Burge, C. B. Science 338, 1593–1599 (2012).
Lambrechts, D. et al. Nat. Med. 24, 1277–1289 (2018).
Rosenberg, A. B. et al. Science 360, 176–182 (2018).
Alavi, A., Ruffalo, M., Parvangada, A., Huang, Z. & Bar-Joseph, Z. Nat. Commun. 9, 4768 (2018).
Zou, H. & Hastie, T. J. R. Stat. Soc. Ser. B. 67, 301–320 (2005).
Carlson, M., Falcon, S., Pages, H. & Li, N. AnnotationDbi: Annotation Database Interface. R package v.1.44.0 (2018).
Jones, K. S. J. Doc. 28, 11–21 (1972).
Baglama, J., Reichel, L. & Lewis, B. W. irlba: Fast Truncated Singular Value Decomposition and Principal Components Analysis for Large Dense and Sparse Matrices. R package v.2.3.3 (2017).
Friedman, J., Hastie, T. & Tibshirani, R. J. Stat. Software 33, 1–22 (2010).
Lun, A. et al. Genome Biol. 20, 63 (2019).
Griffiths, J. A., Richard, A. C., Bach, K., Lun, A. T. L. & Marioni, J. C. Nat. Commun. 9, 2667 (2018).
Acknowledgements
We gratefully acknowledge S. Tapscott, W. Noble and D. Witten as well as members of the Shendure and Trapnell laboratories, particularly A. Hill, for their advice. Z. Pliner named the software. This work was supported by the following funding: NIH grant nos. U54DK107979 and U54HL145611 to J.S. and C.T.; NIH grant nos. DP2HD088158, RC2DK114777 and R01HL118342 to C.T.; NIH grant nos. DP1HG007811 and R01HG006283 to J.S. and the Paul G. Allen Frontiers Group to J.S. and C.T. J.S. is an Investigator of the Howard Hughes Medical Institute. C.T. is partly supported by an Alfred P. Sloan Foundation Research Fellowship. H.A.P. was supported by the National Science Foundation Graduate Research Fellowship under grant no. DGE-1256082.
Author information
Authors and Affiliations
Contributions
C.T. and J.S. conceived the project. H.A.P. wrote Garnett and led the data analysis. H.A.P., C.T. and J.S. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information: Nicole Rusk was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Garnett accurately classifies peripheral blood mononuclear cells.
A) PBMC marker quality plot for 10x version 1 (V1) PBMC dataset (n = 94,571 cells). X-axis corresponds to the ambiguity score, defined as the ratio of the number of ambiguous cells when the marker is included over the number of cells in which the marker is expressed. Color represents the percent of nominated cells for that cell type that were nominated by that marker, and the number next to the cell type names is the total number of nominated cells in that cell type. Markers with an ambiguity score greater than 0.5 (indicated by the red line) were excluded from the marker file. B) Boxplots of the number of unique molecular indexes (UMIs) per cell in 10x Genomics V1 PBMC dataset (n = 94,571 cells) versus version 2 (V2) (n = 8,381 cells) (Boxplot elements: center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, outliers). C) t-SNE plots of 10x Genomics V2 PBMC dataset (n cells = 8,381). Color represents the relative expression of marker genes for each expected cell type (FCGR3A: NK cells, IRF9: Dendritic cells, CD3D: T cells, CD14: Monocytes, CD19: B cells, CD8A and CD8B: CD8 T cells, CD4: CD4 T cells). D) Correspondence between markers of interest and cell clusters in 10x Genomics V2 PBMC dataset with manually assigned cell type labels. Heatmap of relative expression, rows are marker genes and columns are cells sorted by t-SNE cluster assignment. E) t-SNE plot of Garnett cluster-agnostic and cluster-extended type assignments for 10x Genomics V2 PBMCs, also trained on V2 (n = 8,381 cells). F) t-SNE plots of 10x Genomics V2 chemistry applied to PBMCs from a healthy donor (n = 8,381 cells). The first panel is colored by type determined manually using known gene markers. The second and third panels are colored by Garnett cluster-agnostic and cluster-extended cell type assignments by a classifier trained on the 10x Genomics V1 PBMC dataset. G) Correspondence of Garnett cluster-agnostic and cluster-extended type assignments with manual marker-based type assignments for data from (F). Color represents the percent of cells of a certain marker-based type labeled each type by Garnett. H) t-SNE plot of Garnett cluster-agnostic and cluster-extended type assignments for 10x Genomics V1 PBMCs, trained on V2 (n = 94,571 cells). I) Correspondence of Garnett cluster-agnostic and cluster-extended type assignments with FACS assignments for data from (H). Color represents the percent of cells of a certain FACS type labeled each type by Garnett.
Supplementary Figure 2 Garnett is robust to missing cell types and definitions.
A) Classification of 10x Genomics PBMC V1 dataset using a classifier trained on 10x Genomics PBMC V2 dataset with a marker file missing the T cell definition. Panels are t-SNE plots of the V1 dataset (n = 94,571 cells). The first panel is colored by cell type based on FACS sorting, the second panel is colored by cluster-agnostic cell type according to Garnett classification, and the third panel is colored by the Garnett cluster-extended type. B) A heatmap of data in (A) comparing the labels based on FACS (rows) with the cluster-agnostic (left) and cluster-extended (right) cell type assignments by Garnett (columns). Color represents the percent of cells of a certain FACS type labeled each type by Garnett. C) t-SNE plot of 10x Genomics PBMC V1 dataset colored by the number of UMIs mapping to FCGR3A in each cell (n = 94,571 cells). Putative NKT cells indicated by black arrow. D) Similar to (A), classification of 10x Genomics PBMC V1 dataset this time using a classifier trained on 10x Genomics PBMC V2 dataset where the T cells were ablated from the dataset (n = 94,571 cells). The first panel is colored by cell type based on FACS sorting, the second panel is colored by cluster-agnostic cell type according to Garnett classification, and the third panel is colored by the Garnett cluster-extended type. E) A heatmap of data in (D) comparing the labels based on FACS (rows) with the cluster-agnostic (left) and cluster-extended (right) cell type assignments by Garnett (columns). Color represents the percent of cells of a certain FACS type labeled each type by Garnett.
Supplementary Figure 3 Garnett is robust to downsampling.
A) Boxplots of the percent of cells in 10x Genomic PBMC V1 dataset (n = 94,571 cells) correctly classified at various levels of downsampling of V2 training data. X axis shows the percent of original reads kept after downsampling. At each downsampling level, 10 random downsampling sets were used. (Boxplot elements: center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, outliers). B) Boxplots of the percent of cells in 10x Genomic PBMC V1 dataset (n = 94,571 cells) correctly classified at various levels of downsampling of the V1 test dataset. Classifier was trained on the whole 10x Genomic V2 dataset. X axis shows the percent of original reads kept after downsampling. At each downsampling level, 10 random downsampling sets were used. (Boxplot elements: center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, outliers). C) Boxplots of the percent of cells in 10x Genomic PBMC V1 dataset (n = 94,571 cells) correctly classified when varying the number of T cells included in the V2 training data. X-axis shows the number of T cells included in the training set. At each tested level, 10 random sets of T cells were included. (Boxplot elements: center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, outliers).
Supplementary Figure 4 Garnett accurately classifies lung cell types from recent mouse cell atlases.
Panels A, C, E, and G are t-SNE plots of Tabula Muris (TM)2 (n = 1,716 cells) and Mouse Cell Atlas (MCA)3 (n = 6,940) lung subsets colored by reported cell type versus Garnett cluster-agnostic and cluster-extended types. Panels B, D, F and H are heatmaps comparing the reported cell types (rows) versus the Garnett cluster-agnostic and cluster-extended types (columns). Color represents the percent of cells of a certain reported type labelled as each type by Garnett. Panels B, D, F and H correspond with A, C, E, and G respectively. Panels A and B are TM data, and were classified using the TM-trained classifier. Panels C and D are MCA data, and were classified using the MCA-trained classifier. Panels E and F are TM data, and were classified using the MCA-trained classifier. Panels G and H are MCA data, and were classified using the TM-trained classifier.
Supplementary Figure 5 Garnett can classify cells from single-cell chromatin accessibility datasets.
A) t-SNE plot of the Cusanovich et al.7 mouse single-cell ATAC-seq atlas (n = 81,173). Garnett used publicly available Cicero10 gene activity scores in place of expression data to classify cell types. The first panel is colored by Cusanovich et al.7 manually assigned cell type labels. The second and third panel are colored by the Garnett cluster-agnostic and cluster-extended types respectively. B) Heatmaps comparing the reported cell types versus the Garnett cluster-agnostic and cluster-extended types. Color represents the percent of cells of a certain reported type labelled as each type by Garnett.
Supplementary Figure 6 Marker quality chart for C. elegans.
X-axis represents the ambiguity score, defined as the ratio of number of ambiguous cells when the marker is included over the number of cells the marker is expressed in. Color represents the percent of nominated cells for that cell type that were nominated by that marker, and the number next to the cell type names is the total number of nominated cells in that cell type. Markers were initially chosen directly from ref. 11. Markers excluded because of high ambiguity are marked with red boxes.
Supplementary Figure 7 Garnett classification results for sci-RNA-seq data from whole L2 stage C. elegans.
A) t-SNE plots of the whole worm dataset11. First panel is colored by published type from ref. 11, second panel colored by the major (top level) Garnett cluster-agnostic classification (n = 42,035 cells). B) Heatmap comparing the reported cell types versus (rows) the Garnett cluster-agnostic cell type (columns). Color represents the percent of cells of a certain reported type labelled as each type by Garnett.
Supplementary Figure 8 Cell type hierarchy constructed for classification of mouse nervous system cells based on the taxonomy described in ref. 12.
Supplementary Figure 9 Garnett classifies major nervous system types in mouse.
A) Table of the percent of cells correctly identified at each of the cell type hierarchy levels shown in Supplementary Fig. 8 and reproduced in (B). Correct indicates that the Garnett classification was identical to the classification at that hierarchy level in ref. 12. Cell type hierarchy constructed for classification of mouse nervous system cells reproduced from Supplementary Fig. 8. Color represents the hierarchy level used in this figure and Supplementary Fig. 10. C) Heatmaps comparing the reported cell types at hierarchy level 1 versus the Garnett cluster-agnostic and cluster-extended types at hierarchy level 1. Color represents the percent of cells of a certain reported type labelled as each type by Garnett. Red highlighting indicates a correct classification (including correct but underclassified). D) Similar to (C) but hierarchy level 2. E) Similar to (C) but hierarchy level 3.
Supplementary Figure 10 Garnett classifies major nervous system types in mouse.
Heatmaps comparing the reported cell types at hierarchy level 4 versus the Garnett cluster-agnostic and cluster-extended types at hierarchy level 4. Color represents the percent of cells of a certain reported type labelled as each type by Garnett. Red highlighting indicates a correct classification (including correct but underclassified).
Supplementary Figure 11 Features chosen by Garnett have higher expression and high specificity than other genes.
A) Boxplots of the log10 of the total number of reads per gene in the mouse nervous system dataset. Genes are grouped by whether they are input markers (from the marker file, n = 267 genes), classifier features (chosen by elastic-net regression as discriminatory among cell types, n = 1239 genes), or remaining genes (n = 26,492 genes). B) Similar to (A) but showing the log10 of the inverse-document-frequency for each of the genes. (Boxplot elements: center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, outliers; gene n is the same as (A)).
Supplementary Figure 12 Garnett classification of single-cell RNA-seq data from lung tumors.
A) t-SNE plots of lung tumor scRNA-seq dataset14 (n = 52,698 cells). First panel is colored by published type from ref. 14, second panel colored by Garnett cluster-agnostic cell type, and third panel colored by Garnett cluster-extended cell type, based on a model trained using this same dataset. B) Heatmaps comparing the reported cell types versus the Garnett cluster-agnostic and cluster-extended cell types from panel A. Color represents the percent of cells of a certain reported type labelled as each type by Garnett. C) Garnett cluster-agnostic results for human lung tumors from ref. 14 classified based on a Garnett classifier trained on lung cells from the Mouse Cell Atlas. t-SNE plots of the human lung tumor dataset (n = 52,698 cells). First panel is colored by published type from ref. 14; second panel colored by the Garnett cluster-agnostic classification. D) Heatmap comparing the reported cell types versus the Garnett cluster-agnostic cell types from panel C. Color represents the percent of cells of a certain reported type labelled as each type by Garnett.
Supplementary information
Supplementary Information
Supplementary Figs. 1–12, Supplementary Table 1 and Supplementary Note.
Supplementary Dataset 1
Garnett marker file used as input for human PBMC classification.
Supplementary Dataset 2
Garnett marker file used as input for mouse lung classification.
Supplementary Dataset 3
Garnett marker file used as input for scATAC-seq mouse atlas classification.
Supplementary Dataset 4
Garnett marker file used as input for C. elegans classification.
Supplementary Dataset 5
Garnett marker file used as input for human lung classification.
Supplementary Dataset 6
Garnett marker file used as input for mouse nervous system classification.
Rights and permissions
About this article
Cite this article
Pliner, H.A., Shendure, J. & Trapnell, C. Supervised classification enables rapid annotation of cell atlases. Nat Methods 16, 983–986 (2019). https://doi.org/10.1038/s41592-019-0535-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-019-0535-3
This article is cited by
-
Gene panel selection for targeted spatial transcriptomics
Genome Biology (2024)
-
A comparison of marker gene selection methods for single-cell RNA sequencing data
Genome Biology (2024)
-
SRT-Server: powering the analysis of spatial transcriptomic data
Genome Medicine (2024)
-
Enabling methanol fixation of pediatric nasal wash during respiratory illness for single cell sequencing in comparison with fresh samples
Pediatric Research (2024)
-
Single cell atlas of kidney cancer endothelial cells reveals distinct expression profiles and phenotypes
BJC Reports (2024)