Abstract
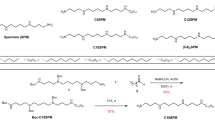
We report the identification of a photocleavable anionic surfactant, 4-hexylphenylazosulfonate (Azo), which can be rapidly degraded by ultraviolet irradiation, for top-down proteomics. Azo can effectively solubilize proteins with performance comparable to that of sodium dodecyl sulfate (SDS) and is compatible with mass spectrometry. Azo-aided top-down proteomics enables the solubilization of membrane proteins for comprehensive characterization of post-translational modifications. Moreover, Azo is simple to synthesize and can be used as a general SDS replacement in SDS–polyacrylamide gel electrophoresis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
All data generated or analyzed during this study are presented in this article or in the provided supplementary materials. Raw gel, blot and mass spectra data are available as Supplementary Data, and source data for Fig. 1 and Supplementary Figs. 4, 9, 10 and 13 are available online. Proteomics data have been uploaded to the PRIDE repository via ProteomeXchange with identifier PXD010825.
References
Aebersold, R. et al. Nat. Chem. Biol. 14, 206–214 (2018).
Siuti, N. & Kelleher, N. L. Nat. Methods 4, 817–821 (2007).
Cai, W. X., Tucholski, T. M., Gregorich, Z. R. & Ge, Y. Expert Rev. Proteomics 13, 717–730 (2016).
Chen, B., Brown, K. A., Lin, Z. & Ge, Y. Anal. Chem. 90, 110–127 (2018).
Barrera, N. P. & Robinson, C. V. Annu. Rev. Biochem 80, 247–271 (2011).
Speers, A. E. & Wu, C. C. Chem. Rev. 107, 3687–3714 (2007).
Loo, R. R., Dales, N. & Andrews, P. C. Protein Sci. 3, 1975–1983 (1994).
Wisniewski, J. R., Zougman, A., Nagaraj, N. & Mann, M. Nat. Methods 6, 359–362 (2009).
Kachuk, C. & Doucette, A. A. J. Proteomics 175, 75–86 (2018).
Chang, Y.-H. et al. J. Proteome Res. 14, 1587–1599 (2015).
Yu, Y. Q., Gilar, M., Lee, P. J., Bouvier, E. S. & Gebler, J. C. Anal. Chem. 75, 6023–6028 (2003).
Chen, E. I., Cociorva, D., Norris, J. L. & Yates, J. R. J. Proteome Res. 6, 2529–2538 (2007).
Meng, F. et al. Anal. Chem. 74, 2923–2929 (2002).
Bradley, M., Vincent, B., Warren, N., Eastoe, J. & Vesperinas, A. Langmuir 22, 101–105 (2006).
Hwang, L., Guardado-Alvarez, T. M., Ayaz-Gunner, S., Ge, Y. & Jin, S. Langmuir 32, 3963–3969 (2016).
Kim, M. S. & Diamond, S. L. Bioorg. Med. Chem. Lett. 16, 4007–4010 (2006).
Dunkin, I. R., Gittinger, A., Sherrington, D. C. & Whittaker, P. J. Chem. Soc. Perkin Trans. I 2, 1837–1842 (1996).
Laganowsky, A., Reading, E., Hopper, J. T. S. & Robinson, C. V. Nat. Protoc. 8, 639–651 (2013).
MacLennan, D. H. & Kranias, E. G. Nat. Rev. Mol. Cell Biol. 4, 566–577 (2003).
He, J. et al. Proc. Natl Acad. Sci. USA 115, 2988–2993 (2018).
Saveliev, S. V. et al. Anal. Chem. 85, 907–914 (2013).
Lee, H. B. et al. J. Org. Chem. 69, 701–713 (2004).
Saveliev, S., Simpson, D. & Wood, K. V. Cleavable surfactants. US patent 0095628 A1 (2009).
Mezger, T., Nuyken, O., Meindl, K. & Wokaun, A. Prog. Org. Coat. 29, 147–157 (1996).
Wientzek, M. & Katz, S. J. Mol. Cell. Cardiol. 23, 1149–1163 (1991).
Peng, Y. et al. Mol. Cell. Proteomics 13, 2752–2764 (2014).
Kou, Q., Xun, L. & Liu, X. Bioinformatics 32, 3495–3497 (2016).
Apweiler, R. et al. Nucleic Acids Res. 32, D115–D119 (2004).
Cai, W. et al. Mol. Cell. Proteomics 15, 703–714 (2016).
Fellers, R. T. et al. Proteomics 15, 1235–1238 (2015).
Ashburner, M. et al. Nature Genet. 25, 25–29 (2000).
Szklarczyk, D. et al. Nucleic Acids Res. 45, D362–D368 (2017).
Cai, W. et al. Anal. Chem. 89, 5467–5475 (2017).
Acknowledgements
This research is supported by National Institutes of Health R01 GM117058 (to S.J. and Y.G.). Y.G. acknowledges R01 HL109810, R01 HL096971, R01 GM125085 and S10 OD018475. MaSDeS was a gift from S. Saveliev (Promega Corporation). We thank A. Chen, E. Chang and W. Tang for their assistance in the early stage of the project, S. Mitchell and T. Tucholski for the help with graphics, and T. Hacker for providing the swine hearts. We thank M. Willetts at Bruker for his assistance with DataAnalysis software. We also acknowledge A. Carr, E. Bayne and J. Melby for their help testing the Supplementary Protocol to ensure reproducible results.
Author information
Authors and Affiliations
Contributions
K.A.B. designed and performed experiments, analyzed the data and wrote the manuscript. B.C. designed and performed experiments, analyzed the data and wrote the manuscript. T.M.G.-A. designed and performed experiments, analyzed the data and wrote the manuscript. Z.L. performed experiments and analyzed the data. L.H. performed experiments and analyzed the data. S.A.-G. performed experiments and analyzed the data. S.J. designed the experiments, supervised the project and wrote the manuscript. Y.G. conceived the idea, designed the experiments, supervised the project and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The University of Wisconsin–Madison has filed a provisional patent application P180335US01, US serial number 62/682027 (7 June 2018) on the basis of this work. Y.G., S.J., K.B. and T.M.G.-A. are named as inventors on the provisional patent application.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–19, Supplementary Table 1 and Supplementary Notes 1–6
Supplementary Table 2
Detected proteoforms from LC–MS/MS run 1 (cardiac tissue)
Supplementary Table 3
Total detected proteoforms in 3 LC–MS/MS experiments: run 1 (cardiac tissue), run 2 (sarcoplasmic reticulum (SR) and mitochondria (Mit) enriched cardiac tissue), and run 3 (SR and Mit enriched cardiac tissue)
Supplementary Table 4
Proteoforms identified by TopPIC from cardiac tissue using Azo from all three LC–MS/MS runs in this study
Supplementary Table 5
Proteins identified by TopPIC from cardiac tissue using Azo from all three LC–MS/MS runs in this study
Supplementary Table 6
Subunits of the electron transport chain identified from cardiac tissue samples using Azo in this study
Supplementary Table 7
Integral membrane proteins identified from cardiac tissue samples using Azo in this study
Supplementary Table 8
ATP synthase subunits identified from cardiac tissue using Azo in this study
Supplementary Table 9
TopPIC proteoform identification: Azo protein extraction of human embryonic kidney (HEK) cell
Supplementary Table 10
TopPIC protein identification: Azo protein extraction of human embryonic kidney (HEK) cell
Supplementary Data
Full gels, blots and raw mass spectra
Rights and permissions
About this article
Cite this article
Brown, K.A., Chen, B., Guardado-Alvarez, T.M. et al. A photocleavable surfactant for top-down proteomics. Nat Methods 16, 417–420 (2019). https://doi.org/10.1038/s41592-019-0391-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-019-0391-1
This article is cited by
-
Therapy-induced senescent tumor cell-derived extracellular vesicles promote colorectal cancer progression through SERPINE1-mediated NF-κB p65 nuclear translocation
Molecular Cancer (2024)
-
Structure and dynamics of endogenous cardiac troponin complex in human heart tissue captured by native nanoproteomics
Nature Communications (2023)
-
Emergence of mass spectrometry detergents for membrane proteomics
Analytical and Bioanalytical Chemistry (2023)
-
Airway fibrin formation cascade in allergic asthma exacerbation: implications for inflammation and remodeling
Clinical Proteomics (2022)
-
FLASHIda enables intelligent data acquisition for top–down proteomics to boost proteoform identification counts
Nature Communications (2022)