Abstract
The derivation of astrocytes from human pluripotent stem cells is currently slow and inefficient. We demonstrate that overexpression of the transcription factors SOX9 and NFIB in human pluripotent stem cells rapidly and efficiently yields homogeneous populations of induced astrocytes. In our study these cells exhibited molecular and functional properties resembling those of adult human astrocytes and were deemed suitable for disease modeling. Our method provides new possibilities for the study of human astrocytes in health and disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Change history
04 December 2018
In the version of Supplementary Fig. 1 originally published with this paper, some images in panel e were accidental duplicates of images in panel b. This error has been corrected in the online integrated supplementary information and in the Supplementary Information PDF.
References
Verkhratsky, A. & Nedergaard, M. Physiol. Rev. 98, 239–389 (2018).
Zhang, Y. et al. Neuron 89, 37–53 (2016).
Oberheim, N. A. et al. J. Neurosci. 29, 3276–3287 (2009).
Caiazzo, M. et al. Stem Cell Rep. 4, 25–36 (2015).
Zamanian, J. L. et al. J. Neurosci. 32, 6391–6410 (2012).
Sofroniew, M. V. & Vinters, H. V. Acta Neuropathol. 119, 7–35 (2010).
Rothstein, J. D. et al. Neuron 16, 675–686 (1996).
Houades, V., Koulakoff, A., Ezan, P., Seif, I. & Giaume, C. J. Neurosci. 28, 5207–5217 (2008).
Newman, E. A. & Zahs, K. R. Science 275, 844–847 (1997).
Parpura, V. et al. Nature 369, 744–747 (1994).
Colombo, E. & Farina, C. Trends Immunol. 37, 608–620 (2016).
Harikumar, K. B. et al. Nat. Immunol. 15, 231–238 (2014).
Chung, W. S., Allen, N. J. & Eroglu, C. Cold Spring Harb. Perspect. Biol. 7, a020370 (2015).
Zhang, Y. et al. Neuron 78, 785–798 (2013).
Tomokane, N., Iwaki, T., Tateishi, J., Iwaki, A. & Goldman, J. E. Am. J. Pathol. 138, 875–885 (1991).
Brenner, M. et al. Nat. Genet. 27, 117–120 (2001).
Chen, M. H., Hagemann, T. L., Quinlan, R. A., Messing, A. & Perng, M. D. ASN Neuro 5, e00125 (2013).
Tian, E. et al. Cell Rep. 16, 781–792 (2016).
Minkel, H. R., Anwer, T. Z., Arps, K. M., Brenner, M. & Olsen, M. L. Glia 63, 2285–2297 (2015).
Saito, K. et al. Glia 66, 1053–1067 (2018).
Canals, I. et al. Protoc. Exch. https://doi.org/10.1038/protex.2018.088 (2018).
Rönn, R. E. et al. Stem Cell Rep. 4, 269–281 (2015).
Yang, N. et al. Nat. Methods 14, 621–628 (2017).
Rothman, J. S. & Silver, R. A. Front. Neuroinform. 12, 14 (2018).
Oki, K. et al. Stem Cells 30, 1120–1133 (2012).
Acknowledgements
We thank L. Oburoglu, N. Woods (Lund Stem Cell Center), and the hES/iPS core facility for providing hiPSCs and technical assistance; C. Zhang (University of Texas Southwestern Medical Center) for the GFAP::GFP vector; T. Velasco-Hernández (Lund Stem Cell Center) and members of the FACS core facility for assistance with cell sorting; E. Erlandsson (Lund Stem Cell Center) and the single-cell genomics facility for assistance with Fluidigm experiments; the StemTherapy Imaging Platform for technical assistance; and the Laboratory of Stem Cells and Restorative Neurology for sharing reagents and for helpful discussions. This study was supported by Foundation Sävstaholm (grant no. ST 2017-008 to E.Q.), the Swedish Research Council (grant no. 521-2014-3541 to H.A.), the Swedish Society for Medical Research (H.A.), the Swedish government initiative for strategic research areas (StemTherapy; H.A.), the Åke Wibergs Foundation (grant no. m17-0100 to H.A.), the Crafoord Foundation (grant no. 20160963 to H.A.), Alzheimerfonden (AF-742791 to H.A.), the King Gustav V and Queen Victoria’s Foundation of Freemasons (grant. no. 20161208 to H.A.), and the Hedlund Family Fund (grant no. M-2017-0557 to H.A.).
Author information
Authors and Affiliations
Contributions
I.C. was responsible for research design, execution, data analysis, and manuscript preparation; I.C., R.T., A.G., E.Q., J.F., I.H., M.G.H., and E.M. performed experiments; G.M. and E.M. provided human astrocytes; D.B. contributed to data analysis and manuscript preparation; J.B. provided clinical resection material and contributed to manuscript preparation; and H.A. was responsible for conceptual and research design, supervision, data interpretation, and manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Screening of transcription factors for astrocyte induction.
(a) Experimental design of screening experiments in hESCs. (b) Representative GFP-fluorescence images for different combinations of Sox9, Nfia and Nfib at 21 days of induction using GFAP::GFP lentivirus. Experiments were performed 3 independent times showing similar results. (c) Representative GFAP::EGFP-fluorescence images of iAs-B and iAs-SB at 7, 14 and 21 days after induction and representative brightfield images at 21 days. Boxes show zoom-in on cells indicated by arrows. Experiments were performed at least 3 independent times showing similar results. (d) Representative phalloidin labeled cells for all factor combinations at 21 days of induction. Experiments were performed 3 independent times showing similar results. (e) Representative images of S100B labeled cells for all factor combinations at 21 days of induction. Experiments were performed 3 independent times showing similar results. B = Nfib, SB = Sox9 + Nfib, d = days. Scale bar = 100 μm.
Supplementary Figure 2 Screening of factors for astrocyte induction, using RT-qPCR and GFAP expression during the first week of induction.
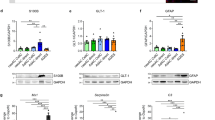
(a) RT-qPCR showing expression of GFAP, S100B and ALDH1L1 after 21 days of induction for all combinations of Nfia, Nfib and Sox9. Results are the mean ± SEM of three independent experiments (GFAP) or the mean of two independent experiments (S100B and ALDH1L1) and are presented as relative expression of the specific gene normalized to GAPDH expression. (b) Quantification of GFAP protein expression by immunolabeling at indicated time points during the first week of induction. Percentage of cells positive for GFAP is shown as mean ± SEM of three independent experiments.
Supplementary Figure 3 iAs-B induction and FACS analysis of iAs-B and iAs-SB populations.
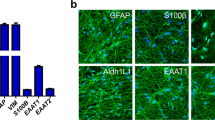
(a) Representative images of iAs-B immunolabeled for indicated astrocytic markers. Experiments were performed 3 independent times showing similar results. (b) Gating strategy used for FACS analyses. Left: forward vs side scatter. Middle: Singlets selection. Right: Viable cells based on 7-AAD staining. (c) Contour plots showing CD44 expression. (d) Contour plots showing VIM expression. Results are shown as mean ± SEM from three independent experiments. 7d = 7 days, 21d = 21 days. Scale bar = 100 μm.
Supplementary Figure 4 iAs morphology, proliferation and glycogen granules.
(a) Representative image of Ki67 stainings for iAs-B and iAs-SB with positive cells marked with an arrow. Experiments were performed 3 independent times showing similar results. (b) Representative immunolabeling of specific astrocytic markers as well as phalloidin at 21 days of induction in iAs-B. Experiments were performed 3 independent times showing similar results. (c) Representative image of glycogen granules staining of iAs-B, SB, human primary astrocytes (hAs) and mouse astrocytes (mAs) and quantification of iAs displaying glycogen granules. Results are shown as mean ± SEM from three independent experiments. (d) Representative images showing immunolabeling for indicated astrocytic markers as well as phalloidin at 2 months after induction. Experiments were performed 3 independent times showing similar results. Scale bar = 25 μm (a), 100 μm (b,d), 10 μm (c).
Supplementary Figure 5 Immunocytochemistry controls.
Representative images for secondary antibody control stainings, rtTA-infected cells, primary astrocytes and iAs-B or iAs-SB labeled cells to detect NSC (PAX6 and SOX2), neurons (MAP2), oligodendrocytes (MBP), microglia (Iba1), renal epithelial cells (ANP) or skeletal muscle cells (MyoD). All images correspond to 21 day induced cells except for primary controls, which correspond to human primary astrocytes in culture. Experiments were performed 3 independent times showing similar results. Scale bar = 100 μm.
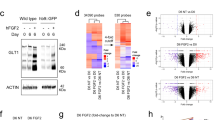
Supplementary Figure 6 Induction of reprogramming factors and Dox dependence.
(a) Expression of human (endogenous) and mouse (exogenous) Nfib and Sox9 at days 0, 7, 14 and 21 with Dox-withdrawal at 7 or 14 days. Results are the mean ± SEM of three independent experiments and presented as relative expression normalized to GAPDH. (b) Representative images of iAs cultures labeled for GFAP and VIM after 21 days of induction with Dox withdrawal at 7 or 14 days. Experiments were performed 3 independent times showing similar results. (c) Quantification of percentage positive cells for GFAP, S100B and VIM at 21 days of induction with Dox withdrawal at 7 or 14 days. Results are mean ± SEM of three independent experiments and presented as percentage of positive cells normalized to the percentage without Dox withdrawal. An ordindary one-way ANOVA with Tukey test correction for multiple comparisons was used to detect significant differences. *P value < 0.05. Scale bar = 100 μm.
Supplementary Figure 7 Characterization of iAs derived from H9 hESC and hiPSC lines.
Representative images showing immunolabeling for the astrocytic markers GFAP, S100B, GLAST and ALDH1L1 as well as phalloidin at 21 days of induction in iAs-B and iAs-SB. Experiments were performed 3 independent times showing similar results. Scale bar = 100 μm.
Supplementary Figure 8 Real-Time qPCR analysis of iAs.
Comparison of expression of different astrocytic genes between iAs-B and iAs-SB at different time points and human adult and embryonic astrocytes. Results are the mean ± SEM of three independent experiments and are presented as relative expression of the specific gene normalized to GAPDH expression.
Supplementary Figure 9 iAs display a homogeneous gene expression signature similar to that of adult human astrocytes.
Single-cell quantitative RT-qPCR analysis (Fluidigm) of the expression of (a) house-keeping and induction, (b) astrocytic, (c) stem cell, oligodendrocytic and neuronal genes. Expression levels are presented in Ct values and color-coded as shown on the top left. Lower Ct values (red) correspond to higher mRNA expression levels. mRNA levels were quantified in single cells FACS-sorted by viability.
Supplementary Figure 10 Functional characterization of iAs.
(a) Representative images showing biocytin spreading in iAS-B at 21 days (left image) and iAs-SB at 21 days (right image). Experiments were performed 3 independent times showing similar results. (b) Representative images showing Fluo-4 fluorescence in iAs-SB 2 seconds (2”) before adding ATP, response 20 seconds (20”) after ATP addition and recovery to basal levels 60 seconds (60”) after ATP stimulation and Fluo-4-based calcium imaging results showing the percentage of cells displaying spontaneous increases in fluorescence in iAs-B, SB and human primary adult astrocyte cultures. Results are the mean ± SEM of three independent experiments. (c) Fold increase in expression of CCL5 and CXCL8 after 8 h of treatment with 10 ng/ml IL-1β for both iAs-B and SB at day 14 and 21 after induction. Results are the mean ± SEM of four independent experiments and are presented as fold increase in expression normalized by GAPDH and relative to the control. Statistical analysis consisted in a one-tailed ratio paired t-test comparing iAs with and without treatment. cyt = IL-1β. *P value < 0.05. **P value < 0.01. ***P value < 0.001. ****P value < 0.0001. (d) Presence of glutamatergic sPSCs is illustrated. Current traces show glutamatergic sPSCs recorded from iNs with mAs (left, black) or with iAs-SB (left, blue) in the presence of 100 μM Ptx and blocked in the presence of 50 μM D-APV + 5 μM NBQX. Expanded current traces illustrate single glutamatergic sPSCs. Bar graph (right) show the frequency of sPSCs in the presence of Ptx and Ptx + D-APV + NBQX in iNs co-cultured with mAs (n = 13 iNs) or with SB-iAs (n = 6 iNs). An unpaired two-tailed Mann-Whitney test was used for statistical analysis. *P value < 0.5 (e) Averaged glutamatergic sPSC detected from recordings in iNs co-cultured with mAs (black, n = 39 sPSCs) or iAs-SB (blue, n = 60 sPSCs) (top). Amplitude of averaged glutamatergic sPSCs from 6 (SB) or 13 (mAs) iNs are illustrated in the bar graph (bottom, p = 0.7813). (f) Pie chart (left) illustrating the amount of iNs with burst-like activity when co-cultured with mAs (black), iAs-SB (blue) or without glia (grey). Current traces from iNs co-cultured with mAs or iAs-SB containing burst-like events and the lack of burst-like events in current traces from iNs without glia (right). (g) Representative images of colocalization analyses for PSD95, Bassoon and MAP2 staining of iNs without glia (No glia) or in co-culture with iAs-B, iAs-SB or mAs. Experiment was performed two independent times with 3 (mAs) or 4 (No glia, B and SB) biological replicates Scale bar = 25 μm (a), 10 μm (b) and 5 μm (g).
Supplementary Figure 11 Characterization of iAs transplanted into mouse brains.
(a) Experimental design for transplantation of 8 neonatal and 4 adult mice. (b) Representative image showing iAs produced in parallel to those used for transplantation and kept in culture until the day when mice were sacrificed. Experiments were performed 3 independent times showing similar results. (c) Representative images of grafts of transplanted iAs labeled for human specific nuclear antigen (HuNu) in neonatal or adult mouse cortex and striatum at 4-5 weeks. Experiments were performed in 4 neonatal and 4 adult transplanted mice showing similar results. (d) Representative image showing GFP + iAs co-labeled with VIM in adult transplanted brains. Experiments were performed in 4 neonatal and 4 adult transplanted mice showing similar results. (e) Representative images of GFP + iAs co-labeled with S100B in neonatal (upper image) and adult (lower image) transplanted mice. Experiments were performed in 4 neonatal and 4 adult transplanted mice showing similar results. (f) Representative orthogonal projections of GFP + iAs co-labeled with GLAST in neonatal (upper image) and adult (lower image) transplanted mice. Experiments were performed in 6 neonatal and 4 adult transplanted mice showing similar results. (g) Representative images of GFP + cells transplanted in neonatal (upper image) or adult (lower image) mice in close interaction with Bassoon + /PSD95 + synaptic puncta. Experiments were performed in 4 neonatal and 3 adult transplanted mice showing similar results. (h) Co-labelling of a GFP positive cell with antibodies for specific astrocytic protein AQP4 and for endothelial specific protein CD31 in an adult transplanted mouse. Experiments were performed in 4 neonatal and 4 adult transplanted mice showing similar results. (i,j) Representative images of Cx43 staining in GFP labeled cells (left image) with orthogonal reconstruction (right image) in a neonatal and adult transplanted mouse. Experiments were performed in 6 neonatal and 4 adult transplanted mice showing similar results. (k,l) Co-labelling of GFP positive transplanted cells filled with biocytin 6 weeks after transplantation (k) and spreading to GFP + /HuNu + transplanted as well as GFP-/HuNu- host cells 13 weeks after transplantation (l). Magenta arrow marks injected cell, green and yellow arrows mark transplanted and host cells, respectively, with spreading of biocytin. Experiments were performed 4 independent times showing similar results. Scale bar = 100 μm (b), 200 μm (c), 10 μm (d,e,g,h), 5 μm (f,i,j) and 20 μm (k,l). LV = lateral ventricle.
Supplementary Figure 12 Genome-engineering strategy for AxD, absence of off-target editing, and characterization of AxD iAs.
(a) Vector construct (left) encoding Cas9n and sgRNAs 1 and 2 targeting regions near the R239C mutation site in exon 4 of GFAP (underlined). Repair template (ssDNA) that introduced the AxD mutation (in red) and a silent change in one PAM sequence (in green) to avoid repetitive editing (middle). Sequencing results of one homozygous clone obtained showing both changes in the genome (right). (b) Chromatograms of genomic regions in chromosomes (Chr) with possible target sequences similar to sgRNAs A and B (marked in blue). Chromatograms include 40 nucleotides up-stream and down-stream of the target sequence. Reference sequences (Ref Seq) are in bold in the first row of each section followed by the WT chromatogram and the AxD chromatogram. Red box in Chr10 for sgRNA B correspond to a described polymorphism (rs74111799) present also in the isogenic WT. (c) Expression of S100B at 21 days of differentiation for WT- and AxD-iAs. Results are the mean ± SEM of three independent experiments and are presented as relative expression of the specific gene normalized by GAPDH expression. (d) GFAP and S100B labeling of iAs-SB at 28 days after induction for the WT and the AxD lines. Arrows mark cells with GFAP inclusions. Experiments were performed 2 independent times showing similar results. (e) Expression of KCNJ10 at 14 and 21 days of differentiation for WT- and AxD-iAs. Results are the mean ± SEM of three independent experiments and are presented as relative expression normalized by GAPDH expression. An unpaired two-tailed t-test with Welch’s correction for unequal variances was performed. *P value < 0.05. (f) Percentage of cells responding to ATP in calcium imaging assays for WT- and AxD-iAs at 21 days of differentiation. Results are the mean ± SEM of three independent experiments. (g) Fluo-4-based calcium imaging results showing the fold increase at the peak of the response (right). Results are the mean ± SEM of 63 (WT) and 51 (AxD) responding cells analyzed from three independent experiments. Box-whisker plot shows from down to top, the minimum, first quartile, median, third quartile and maximum values. An unpaired two-tailed Mann-Whitney test was used to detect significant differences. *P value < 0.05. Scale bar = 50 μm.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12, Supplementary Tables 1–8 and Supplementary Notes 1–4
Supplementary Protocol
Supplementary Protocol
Rights and permissions
About this article
Cite this article
Canals, I., Ginisty, A., Quist, E. et al. Rapid and efficient induction of functional astrocytes from human pluripotent stem cells. Nat Methods 15, 693–696 (2018). https://doi.org/10.1038/s41592-018-0103-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-018-0103-2
This article is cited by
-
Morphological diversification and functional maturation of human astrocytes in glia-enriched cortical organoid transplanted in mouse brain
Nature Biotechnology (2024)
-
Liprin-α proteins are master regulators of human presynapse assembly
Nature Neuroscience (2024)
-
LET-381/FoxF and its target UNC-30/Pitx2 specify and maintain the molecular identity of C. elegans mesodermal glia that regulate motor behavior
The EMBO Journal (2024)
-
Induced Pluripotent Stem Cells as a Possible Approach for Exploring the Pathophysiology of Polycystic Ovary Syndrome (PCOS)
Stem Cell Reviews and Reports (2024)
-
Human Alzheimer’s disease reactive astrocytes exhibit a loss of homeostastic gene expression
Acta Neuropathologica Communications (2023)