Abstract
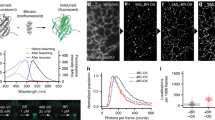
Reversibly photoswitchable fluorescent proteins (rsFPs) are gaining popularity as tags for optical nanoscopy because they make it possible to image with lower light doses. However, green rsFPs need violet-blue light for photoswitching, which is potentially phototoxic and highly scattering. We developed new rsFPs based on FusionRed that are reversibly photoswitchable with green-orange light. The rsFusionReds are bright and exhibit rapid photoswitching, thereby enabling nanoscale imaging of living cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Hell, S. W. Science 316, 1153–1158 (2007).
Hell, S. W., Jakobs, S. & Kastrup, L. Appl. Phys. A 77, 859–860 (2003).
Grotjohann, T. et al. Nature 478, 204–208 (2011).
Shcherbakova, D. M., Sengupta, P., Lippincott-Schwartz, J. & Verkhusha, V. V. Annu. Rev. Biophys. 43, 303–329 (2014).
Brakemann, T. et al. Nat. Biotechnol. 29, 942–947 (2011).
Grotjohann, T. et al. eLife 1, e00248 (2012).
Duwé, S. et al. ACS Nano 9, 9528–9541 (2015).
Wang, S. et al. ACS Nano 10, 9136–9144 (2016).
Tiwari, D. K. et al. Nat. Methods 12, 515–518 (2015).
Testa, I. et al. Neuron 75, 992–1000 (2012).
Habuchi, S. et al. Proc. Natl Acad. Sci. USA 102, 9511–9516 (2005).
Rego, E. H. et al. Proc. Natl Acad. Sci. USA 109, E135–E143 (2012).
Dedecker, P., Mo, G. C., Dertinger, T. & Zhang, J. Proc. Natl Acad. Sci. USA 109, 10909–10914 (2012).
Chudakov, D. M. et al. Nat. Biotechnol. 21, 191–194 (2003).
Stiel, A. C. et al. Biophys. J. 95, 2989–2997 (2008).
Subach, F. V. et al. Chem. Biol. 17, 745–755 (2010).
Hofmann, M., Eggeling, C., Jakobs, S. & Hell, S. W. Proc. Natl Acad. Sci. USA 102, 17565–17569 (2005).
Lavoie-Cardinal, F. et al. Chemphyschem 15, 655–663 (2014).
Shemiakina, I. I. et al. Nat. Commun. 3, 1204 (2012).
Klementieva, N. V. et al. Chem. Commun. (Camb.) 53, 949–951 (2017).
Pletnev, S., Subach, F. V., Dauter, Z., Wlodawer, A. & Verkhusha, V. V. J. Mol. Biol. 417, 144–151 (2012).
Masullo, L. et al. bioRxiv Preprint at https://www.biorxiv.org/content/early/2018/01/17/248880 (2018).
Chmyrov, A. et al. Nat. Methods 10, 737–740 (2013).
Mishina, N. M. et al. Nano Lett. 15, 2928–2932 (2015).
Schell, M. J., Erneux, C. & Irvine, R. F. J. Biol. Chem. 276, 37537–37546 (2001).
Higuchi, Y., Ueda, Y., Ogawa, H. & Saiki, J. Atten. Percept. Psychophys. 78, 2397–2410 (2016).
Vartanian, J. P., Henry, M. & Wain-Hobson, S. Nucleic Acids Res. 24, 2627–2631 (1996).
Bindels, D. S. et al. Nat. Methods 14, 53–56 (2017).
Merzlyak, E. M. et al. Nat. Methods 4, 555–557 (2007).
Acknowledgements
We thank E. Uriati for help in recording data for the phototoxicity experiments. rsCherryRev1.4–vimentin and rsCherryRev–actinChromobody plasmids were a kind gift from S. Hell and S. Jakobs (Max Planck Institute for Biophysical Chemistry, Göttingen, Germany). This work was supported by ERC starting grant MoNaLISA (https://doi.org/10.13039/501100000781; to I.T.), the Russian Science Foundation (grant 16-14-10364 to A.S.M. and I.I.S.), the US National Institutes of Health (grants GM122567 and NS103573 to V.V.V.), and the Brazilian CNPq (to A.R.F.). J.D. acknowledges the Carlsberg Foundation for support. Experiments were partially carried out with equipment provided by the CKP IBCH core facility, which is supported by Russian Ministry of Education and Science grant RFMEFI62117X0018.
Author information
Authors and Affiliations
Contributions
I.T. designed the project and the imaging platform. V.V.V., K.A.L., D.M.C., and A.S.M. conceived and engineered the new rsFusionRed variants. I.T. and F.P. built the setup and performed the photoswitching and imaging. I.I.S. and A.R.F. performed mutagenesis and screening. E.O.S., I.I.S., A.R.F., N.G.B., A.A.K., and A.S.M. characterized the proteins. N.G.B. and N.G.G. did the cloning. J.D. performed the point-scanning experiments, and A.B. designed the software. I.T. and A.S.M. wrote the manuscript with assistance from all of the other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Alignment of rsFusionRed1, -2, and -3 sequences with those of other red photoswitchable fluorescent proteins.
Sequence of avGFP is shown for numbering, buried residues are shaded in grey. Substitutions important for photoswitching properties (in comparison with non-photoswitchable predecessor) are highlighted in black.
Supplementary Figure 2 Monomeric nature of rsFusionRed.
Size-exclusion chromatography of rsFusionRed variants. A protein probe (~100 µg) was applied to a Superdex 200 column (Phenomenex) connected to a Shimadzu chromatography system (Shimadzu, Japan). Size-exclusion chromatography of samples was performed by constant flow rate 0.4 mL/min of 50 mM Tris-HCl, pH8.0, supplied with 150 mM NaCl. Chromatographic profiles with absorption at 585 nm were detected and overlaid for further analysis. Experiments were repeated twice on independently expressed and purified protein samples with similar results. Curves from one experiment are shown. rsFusionRed are indistinguishable from FusionRed or mCherry, confirming their monomeric state.
Supplementary Figure 3 Spectral properties of the rsFusionReds.
Normalized absorbance spectra are shown for freshly purified (black lines) proteins and the spectra immediately after 5 min of illumination with red LED (590 nm, ~1 mW/cm2). Bottom panel displays the normalized fluorescence excitation (dashed lines) and emission (solid lines) spectra. Shown are the representative curves from independently expressed and purified protein samples (n = 3).
Supplementary Figure 4 Quantum yield estimation of rsFusionReds.
The fluorescence quantum yield of the protein in thermal equilibrium (21 °C) was estimated by fitting the absorbance vs. area of fluorescence data to a linear model and comparing the slope with the reference (mCherry, QY = 0.231). 1Bindels, D.S. et al. mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nature methods 14, 53-56 (2017).
Supplementary Figure 5 Influence of the ON-switching wavelength for rsFusionRed3.
(a) ON-switching efficiency for 405 nm, 488 nm and 510 nm illumination at equal 590 nm read-out light (0.18 kW/cm2 for 100 ms, corresponding to the complete switching into the OFF state for rsFusionRed3). The curves have been fitted with a mono-exponential (solid line); error bars indicate ± s.d over the mediated area of the cell. No significant difference is reported between 405 nm and 488 nm illumination, while 510 nm light is slightly slower (1.5 times) but reaches the same plateau. (b) Switching fatigue related to the different ON-switching illumination. The ON-switching light doses selected for the different wavelengths correspond to 80% of rsFusioRed into the ON state (1.15 J/cm2 for both 405 and 488 nm and 2.7 J/cm2 of 510 nm) highlighted by the red dashed line in graph (a). The measurements have been repeated twice with similar results.
Supplementary Figure 6 Switching fatigue curves at different ON-switching illumination wavelengths.
Given a fixed read-out illumination of 0.18 kW/cm2 at 590 nm, we analysed the fatigue resistance of the different rsFusionRed at the two wavelengths of ON-switching, 405 nm (dark color line) and 510 nm (light color line). According to the ON-switching curves of Fig. 1d, the intensity needed, to recruit 80% of the molecule to the ON state, has been used for the comparison of the fatigue resistance, corresponding to 2.2 J/cm2 of 405 nm and 2.6 J/cm2 of 510 nm for rsFusionRed1; 1.64 J/cm2 of 405 nm and 3.2 J/cm2 of 510 nm for rsFusionRed2; 1.5 J/cm2 of 405 nm and 2.1 J/cm2 of 510 nm for rsFusionRed3. The measurements have been repeated twice with similar results.
Supplementary Figure 7 Thermic return to the ON state.
The whole cell has been exposed to the 590 nm laser light at 0.02 kW/cm2 until the complete switching OFF of the fluorescence signal and the subsequent fluorescence recovery has been probed at different time points for an hour. Each trace is normalized to the fluorescence level prior to complete OFF-switching. All the experiments are performed in live U2OS cells expressing the redFPs constructs rsFusionRed1-vimentin, rsFusionRed2-vimentin and rsFusionRed3-vimentin respectively. The time trace for the different proteins shows a recovery of half of the initial fluorescence after 22 min for rsFusionRed1, 37 min for rsFusionRed2 and 43 min for rsFusionRed3. The measurements have been repeated twice with similar results.
Supplementary Figure 8 Optical scheme of MoNaLISA nanoscopy for imaging of red rsFPs.
The optical elements drawn with dashed lines represent removable elements.
Supplementary Figure 9 MoNaLISA imaging with light above 510 nm.
We performed ON-switching with green light at 510 nm and OFF-switching and read-out with orange light at 590 nm of rsFusionRed3-F-Tractin labelled cells. Scale bar, 5 μm. As before, we could resolve structures smaller than 80 nm (ROI I, scale bar, 500 nm) and (b) follow them over time for more than 25 sub-resolved frames (ROI II, scale bar, 500 nm). The images and data presented are representative of at least 3 experiments with consistent results.
Supplementary Figure 10 Light-induced DNA damage under different imaging conditions for different rsFPs.
Graph (a) shows the light-induced DNA repair puncta increase for different wavelengths and light doses needed for different rsFPs imaging. We plotted the ratio of the number of puncta measured in the nuclei of cells recorded after MoNaLISA imaging divided by the number measured in the initial frame before super-resolution imaging. We repeated the experiments for illumination intensities typical of our rsFusionRed (green, n = 13), rsEGFP2 imaging (violet, n = 10) and rsCheeryRev1.4 (red, n = 17). The control (grey, n = 10) is the same ratio with no MoNaLISA recording. The colored boxes and bars represent the extension of 25% and 95% of the distribution respectively. The x represent statistical outliers. The average and median of the distribution are represented by a square and a line respectively. Graph (b) shows the same ratio after 10 times longer MoNaLISA imaging with 405 nm (violet, n = 10) and 510 nm (green, n = 11) ON-switching. (c) Representative images of nuclei (n = 13-17) of U2OS cells expressing XRCC1-EGFP recorded before and after one MoNaLISA imaging. The grey circles are the DNA repair points detected in the control where no MoNaLISA imaging is performed. The green, blue and red circles are the results recorded before and after the MoNALISA illumination pulse scheme for rsFusionRed3, rsEGFP2 and rsCherryRev1.4 respectively. Scale bars, 10 μm.
Supplementary Figure 11 Switching kinetics and RESOLFT imaging with a focused light beam.
(a) We measured a significant difference in the trend of the OFF-kinetics curves depending on the illumination wavelength at 562 nm (upper two graphs) and 592 nm (lower two graphs), thus affecting both background and switching speed. The laser light intensity was 58 kW/cm2 in both experiments. Interestingly, even at relatively low power ( > 1 kW/cm2) the rsFusionRed2 and 3 follow a bi-exponential decay, as can be seen in the inset where rsFusionRed2 was illuminated with 14 kW/cm2 of 592 nm laser light. All curves are the average of the first 30-50 cycles. They were measured on cells expressing either rsFusionRed2-Vimentin or rsFusionRed3-Vimentin. (b) We recorded OFF-switching curves at different light intensities. The plot shows the weighted average of the two half-life components of the bi-exponential fit as a function of the OFF-switching light intensities. As expected the half time decreases with increased 590 nm light intensities for rsFusionRed2, rsFusionRed3 and rsCherryRev1.4, with the latter being about 6 times slower than the rsFusionRed. The results presented for the switching kinetics are the results of a single experiment, but all experiments were performed at least twice. (c) Optical scheme of the point-scanning RESOLFT set-up dedicated to rsFusionReds imaging. (d) RESOLFT images of vimentin structures tagged with rsFuionRed3 compared with conventional images acquired without the OFF-switching step. The images are shown both as raw data and with a Gaussian smooth of 1.2 pixels. The illumination pulse scheme performed pixel-by-pixel was the following: ON-switching with a focused beam at 405 nm, duration 0.5 ms, 24 W/cm2; OFF-switching with a donut-shaped beam at 592 nm, duration 5 ms, 9 kW/cm2; read-out with a focused beam at 560 nm, 0.1 ms duration, 16 kW/cm2. Pixel size 30 nm. The images are representative of at least two experiments with consistent results. Scale bars, 500 nm.
Supplementary Figure 12 Speeding up the rsFP OFF-switching cycle.
The OFF-switching illumination time can be reduced to 2.5 ms without loss in spatial resolution. The amount of OFF-switching illumination photon flux is kept constant by decreasing the size of the illuminated area. Increasing the orange illumination laser power, when available, lead to the same result. (a) Wide-field (inset), enhanced confocal (inset) and MoNaLISA imaging of living U2OS cells expressing rsFusionRed3-F-Tractin. Scale bar, 5 μm. (b) Zoom-in of the white rectangle. (c) Normalized intensity profiles along the line between the white arrowheads illustrated in panel (b) for the enhanced confocal (grey line) and MoNaLISA image (data in black circle, fitted curves in black and orange lines). The images and data presented are representative of at least 2 experiments with consistent results.
Supplementary Figure 13 Vimentin expression and MoNaLISA imaging of red-emitting rsFPs.
Enhanced confocal images of the vimentin filament network of U2OS cells tagged respectively with rsCherryRev1.4 (a) and rsFusionRed3 (b). MoNaLISA images of the same structures are shown in (c) and (d) respectively for rsCherryRev1.4 and rsFusionRed3. The region of interest I is represented in three images recorded in the enhanced confocal mode with no OFF-switching (left), MoNaLISA with 15 ms OFF-switching (middle) and MoNaLISA with 60 ms OFF-switching (right) respectively, featuring a FWHM of 252 nm, 188 nm and 109 nm respectively. The region of interest II shows the enhanced confocal and MoNaLISA (with 15 ms of OFF-switching) images of rsFusionRed3-Vimentin tagged filaments featuring a FWHM of 252 nm and 60 nm respectively. (e) and (f) are the line profiles averaged on 3 adjacent pixels, corresponding to a filament length of 105 nm, and respective Gaussian fits for rsCherryRev1.4 and rsFuionRed3-tagged filaments respectively. The images and data presented are representative of at least 5 experiments with consistent results.
Supplementary Figure 14 In vitro protein maturation.
After expression under low-oxygen conditions, proteins were quickly purified on ice and then left to mature at 37 °C. The fluorescence spectra (excitation 550 nm) were collected at 1 minute intervals. Similar results were obtained in two independent experiments; curves from one experiment are shown.
Supplementary information
Supplementary Text and Figures
Supplementary Figs. 1–14, Supplementary Notes 1 and 2 and Supplementary Tables 1 and 2
Rights and permissions
About this article
Cite this article
Pennacchietti, F., Serebrovskaya, E.O., Faro, A.R. et al. Fast reversibly photoswitching red fluorescent proteins for live-cell RESOLFT nanoscopy. Nat Methods 15, 601–604 (2018). https://doi.org/10.1038/s41592-018-0052-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-018-0052-9
This article is cited by
-
Super-sectioning with multi-sheet reversible saturable optical fluorescence transitions (RESOLFT) microscopy
Nature Methods (2024)
-
Blue-shift photoconversion of near-infrared fluorescent proteins for labeling and tracking in living cells and organisms
Nature Communications (2023)
-
Enhanced detection of fluorescence fluctuations for high-throughput super-resolution imaging
Nature Photonics (2023)
-
Absolute measurement of cellular activities using photochromic single-fluorophore biosensors and intermittent quantification
Nature Communications (2022)
-
First biphotochromic fluorescent protein moxSAASoti stabilized for oxidizing environment
Scientific Reports (2022)