Abstract
Immune-regulatory mechanisms of drug-free remission in rheumatoid arthritis (RA) are unknown. We hypothesized that synovial tissue macrophages (STM), which persist in remission, contribute to joint homeostasis. We used single-cell transcriptomics to profile 32,000 STMs and identified phenotypic changes in patients with early/active RA, treatment-refractory/active RA and RA in sustained remission. Each clinical state was characterized by different frequencies of nine discrete phenotypic clusters within four distinct STM subpopulations with diverse homeostatic, regulatory and inflammatory functions. This cellular atlas, combined with deep-phenotypic, spatial and functional analyses of synovial biopsy fluorescent activated cell sorted STMs, revealed two STM subpopulations (MerTKposTREM2high and MerTKposLYVE1pos) with unique remission transcriptomic signatures enriched in negative regulators of inflammation. These STMs were potent producers of inflammation-resolving lipid mediators and induced the repair response of synovial fibroblasts in vitro. A low proportion of MerTKpos STMs in remission was associated with increased risk of disease flare after treatment cessation. Therapeutic modulation of MerTKpos STM subpopulations could therefore be a potential treatment strategy for RA.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Ranked lists of STM cluster markers, condition-specific markers of MerTKpos STM clusters identified by comparison of two clinical conditions, multiple conditions, STM cluster pathway analysis, FLS cluster markers and DEGs of lining-layer FLS clusters comparing active and remission synovium are provided in the Supplementary Dataset. The list of genes in the custom stromal panel, sample scRNA-seq metrics of FLS cocultured with STM and sequenced with scRNA-seq BD Rhapsody system, and the list of DEGs from FLS–MoM cocultured with MerTK inhibitor, are provided in the Supplementary Dataset. All raw and processed data of STM scRNA-seq, STM–FLS coculture scRNA-seq and MoM–FLS coculture bulk RNA-seq (FLS) were deposited at EMBL-EBI and are available with the following accession numbers: E-MTAB-8322, E-MTAB-8873 and E-MTAB-8316.
Code availability
The Seurat objects and codes used are available from the corresponding authors upon reasonable request.
References
Nagy, G. & van Vollenhoven, R. F. Sustained biologic-free and drug-free remission in rheumatoid arthritis, where are we now? Arthritis Res. Ther. 17, 181 (2015).
Alivernini, S. et al. Tapering and discontinuation of TNF-alpha blockers without disease relapse using ultrasonography as a tool to identify patients with rheumatoid arthritis in clinical and histological remission. Arthritis Res. Ther. 18, 39 (2016).
Baker, K. F. et al. Predicting drug-free remission in rheumatoid arthritis: a prospective interventional cohort study. J. Autoimmun. 105, 102298 (2019).
Alivernini, S. et al. Synovial features of patients with rheumatoid arthritis and psoriatic arthritis in clinical and ultrasound remission differ under anti-TNF therapy: a clue to interpret different chances of relapse after clinical remission? Ann. Rheum. Dis. 76, 1228–1336 (2017).
Rauber, S. et al. Resolution of inflammation by interleukin-9-producing type 2 innate lymphoid cells. Nat. Med. 23, 938–944 (2017).
Firestein, G. S., Gabriel, S. E., McInnes, I. B. & O’Dell, J. R. Kelley and Firestein’s Textbook of Rheumatology 2nd edn (Elsevier, 2016).
Misharin, A. V. et al. Nonclassical Ly6C(–) monocytes drive the development of inflammatory arthritis in mice. Cell Rep. 9, 591–604 (2014).
Gomez Perdiguero, E. et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518, 547–551 (2015).
Culemann, S. et al. Locally renewing resident synovial macrophages provide a protective barrier for the joint. Nature 572, 670–675 (2019).
Mandelin, A. M. 2nd et al. Transcriptional profiling of synovial macrophages using minimally invasive ultrasound-guided synovial biopsies in rheumatoid arthritis. Arthritis Rheumatol. 70, 841–854 (2018).
Kurowska-Stolarska, M. & Alivernini, S. Synovial tissue macrophages: friend or foe? RMD Open 3, e000527 (2017).
Udalova, I. A., Mantovani, A. & Feldmann, M. Macrophage heterogeneity in the context of rheumatoid arthritis. Nat. Rev. Rheumatol. 12, 472–485 (2016).
Herenius, M. M. et al. Monocyte migration to the synovium in rheumatoid arthritis patients treated with adalimumab. Ann. Rheum. Dis. 70, 1160–1162 (2011).
Weiss, M. et al. IRF5 controls both acute and chronic inflammation. Proc. Natl Acad. Sci. USA 112, 11001–11006 (2015).
Yeo, L. et al. Expression of chemokines CXCL4 and CXCL7 by synovial macrophages defines an early stage of rheumatoid arthritis. Ann. Rheum, Dis. 75, 763–771 (2016).
Firestein, G. S. & McInnes, I. B. Immunopathogenesis of rheumatoid arthritis. Immunity 46, 183–196 (2017).
Kurowska-Stolarska, M. et al. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc. Natl Acad. Sci. USA 108, 11193–11198 (2011).
Croft, A. P. et al. Distinct fibroblast subsets drive inflammation and damage in arthritis. Nature. 570, 246–251 (2019).
Zhang, F. et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 20, 928–942 (2019).
Mizoguchi, F. et al. Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat. Commun. 9, 789 (2018).
Stephenson, W. et al. Single-cell RNA-seq of rheumatoid arthritis synovial tissue using low-cost microfluidic instrumentation. Nat. Commun. 9, 791 (2018).
Kuo, D. et al. HBEGF(+) macrophages in rheumatoid arthritis induce fibroblast invasiveness. Sci. Transl. Med. 11, eaau8587 (2019).
Smolen, J. S. et al. Rheumatoid arthritis. Nat. Rev. Dis. Primers 4, 18001 (2018).
Gremese, E. M., Fedele, A. L., Alivernini, S. & Ferraccioli, G. Ultrasound assessment as predictor of disease relapse in children and adults with arthritis in clinical stable remission: new findings but still unmet needs. Ann. Rheum. Dis. 77, 1391–1393 (2018).
Najm, A. et al. Standardisation of synovial biopsy analyses in rheumatic diseases: a consensus of the EULAR Synovitis and OMERACT Synovial Tissue Biopsy Groups. Arthritis Res. Ther. 20, 265 (2018).
Singh, J. A., Arayssi, T., Duray, P. & Schumacher, H. R. Immunohistochemistry of normal human knee synovium: a quantitative study. Ann. Rheum. Dis. 63, 785–790 (2004).
Davies, L. C., Jenkins, S. J., Allen, J. E. & Taylor, P. R. Tissue-resident macrophages. Nat. Immunol. 14, 986–995 (2013).
A-Gonzales, N. et al. Phagocytosis imprints heterogeneity in tissue-resident macrophages. J. Exp. Med. 214, 1281–1296 (2017).
Hogg, N., Palmer, D. G. & Revell, P. A. Mononuclear phagocytes of normal and rheumatoid synovial membrane identified by monoclonal antibodies. Immunology 56, 673–681 (1985).
Bykerk, V. P. & Massarotti, E. M. The new ACR/EULAR remission criteria: rationale for developing new criteria for remission. Rheumatology (Oxford) 51, vi16–vi20 (2012).
Alivernini, S. et al. Synovial predictors of differentiation to definite arthritis in patients with seronegative undifferentiated peripheral inflammatory arthritis: microRNA signature, histological, and ultrasound features. Front. Med. (Lausanne) 5, 186 (2018).
Nolting, J. et al. Retinoic acid can enhance conversion of naive into regulatory T cells independently of secreted cytokines. J. Exp. Med. 206, 2131–2139 (2009).
Vogt, L. et al. VSIG4, a B7 family-related protein, is a negative regulator of T cell activation. J. Clin. Invest. 116, 2817–2826 (2006).
Rothlin, C. V., Ghosh, S., Zuniga, E. I., Oldstone, M. B. & Lemke, G. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell. 131, 1124–1136 (2007).
van der Touw, W., Chen, H. M., Pan, P. Y. & Chen, S. H. LILRB receptor-mediated regulation of myeloid cell maturation and function. Cancer Immunol. Immunother. 66, 1079–1087 (2017).
Kahles, F., Findeisen, H. M. & Bruemmer, D. Osteopontin: a novel regulator at the crossroads of inflammation, obesity and diabetes. Mol. Metab. 3, 384–393 (2014).
Wang, S. et al. S100A8/A9 in Inflammation. Front. Immunol. 9, 1298 (2018).
Lim, H. Y. et al. Hyaluronan receptor LYVE-1-expressing macrophages maintain arterial tone through hyaluronan-mediated regulation of smooth muscle cell collagen. Immunity 49, 326–341 (2018).
Freeman, C. L. et al. Cytokine release in patients with CLL treated with obinutuzumab and possible relationship with infusion-related reactions. Blood 126, 2646–2649 (2015).
Villani, A. C. et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573 (2017).
Boulet, S. et al. The orphan nuclear receptor NR4A3 controls the differentiation of monocyte-derived dendritic cells following microbial stimulation. Proc. Natl Acad. Sci. USA 116, 15150–15159 (2019).
Humby, F. et al. Synovial cellular and molecular signatures stratify clinical response to csDMARD therapy and predict radiographic progression in early rheumatoid arthritis patients. Ann. Rheum. Dis. 78, 761–772 (2019).
Lewis, M. J. et al. Molecular portraits of early rheumatoid arthritis identify clinical and treatment response phenotypes. Cell Rep. 28, 2455–2470 (2019).
Vattakuzhi, Y., Abraham, S. M., Freidin, A., Clark, A. R. & Horwood, N. J. Dual-specificity phosphatase 1-null mice exhibit spontaneous osteolytic disease and enhanced inflammatory osteolysis in experimental arthritis. Arthritis Rheum. 64, 2201–2210 (2012).
Roberts, A. W. et al. Tissue-resident macrophages are locally programmed for silent clearance of apoptotic cells. Immunity 47, 913–927 (2017).
Koenis, D. S. et al. Nuclear receptor Nur77 limits the macrophage inflammatory response through transcriptional reprogramming of mitochondrial metabolism. Cell Rep. 24, 2127–2140 (2018).
Hanna, R. N. et al. NR4A1 (Nur77) deletion polarizes macrophages toward an inflammatory phenotype and increases atherosclerosis. Circ. Res. 110, 416–427 (2012).
Mahajan, S. et al. Nuclear receptor Nr4a2 promotes alternative polarization of macrophages and confers protection in sepsis. J. Biol. Chem. 290, 18304–18314 (2015).
Wood, M. J. et al. Macrophage proliferation distinguishes 2 subgroups of knee osteoarthritis patients. JCI Insight 4, e125325 (2019).
Waterborg, C. E. J. et al. Protective role of the MER tyrosine kinase via efferocytosis in rheumatoid arthritis models. Front. Immunol. 9, 742 (2018).
Jaitin, D. A. et al. Lipid-associated macrophages control metabolic homeostasis in a trem2-dependent manner. Cell. 178, 686–698 (2019).
Boeters, D. M., Burgers, L. E., Toes, R. E. & van der Helm-van Mil, A. Does immunological remission, defined as disappearance of autoantibodies, occur with current treatment strategies? A long-term follow-up study in rheumatoid arthritis patients who achieved sustained DMARD-free status. Ann. Rheum. Dis. 78, 1497–1504 (2019).
Aletaha, D. et al. 2010 rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Ann. Rheum. Dis. 69, 1580–1588 (2010).
Alten, R. et al. Developing a construct to evaluate flares in rheumatoid arthritis: a conceptual report of the OMERACT RA Flare Definition Working Group. J. Rheumatol. 38, 1745–1750 (2011).
Machado, P. et al. Multinational evidence-based recommendations on how to investigate and follow-up undifferentiated peripheral inflammatory arthritis: integrating systematic literature research and expert opinion of a broad international panel of rheumatologists in the 3E Initiative. Ann. Rheum. Dis. 70, 15–24 (2011).
Alivernini, S. et al. MicroRNA-155 influences B-cell function through PU.1 in rheumatoid arthritis. Nat. Commun. 7, 12970 (2016).
Krenn, V. et al. Synovitis score: discrimination between chronic low-grade and high-grade synovitis. Histopathology 49, 358–364 (2006).
Liu, J. et al. UNC1062, a new and potent Mer inhibitor. Eur. J. Med. Chem. 65, 83–93 (2013).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Finak, G. et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015).
Acknowledgements
We thank the patients and healthy volunteers who participated in this study. We thank D. Vaughan for her outstanding support in cell sorting and phenotyping; D. Riggans (University of Glasgow Mail Room), who facilitated smooth deliveries of clinical samples from Rome to Glasgow and Oxford; A. Corbyn, T. Khoyratty and J. Webber (The Kennedy Institute of Rheumatology, Oxford, UK) for assistance in preparing cells for single-cell sequencing; C. Giampà and O. Parolini (Istituto di Anatomia Umana e Biologia Cellulare, Università Cattolica del Sacro Cuore, Rome, Italy) for support with fluorescent microscopy; J. Galbraith and P. Herzyk (Glasgow Polyomics) for rapid scRNA-seq of samples for the revised version of this manuscript; and L. Lemgruber Soares for help with confocal microscopy. This work was supported by the Research into Inflammatory Arthritis Centre Versus Arthritis UK, based in the Universities of Glasgow, Birmingham, Newcastle and Oxford (nos. 20298 and 22072) to L.M., S.F., S.N.S., A.F., A.R.C., I.U., C.D.B., T.D.O., I.B.M. and M.K.-S.; linea D1 (no. R4124500654) (Università Cattolica del Sacro Cuore, no. R4124500654, to S.A.); Ricerca Finalizzata Ministero della Salute (no. GR-2018-12366992, to S.A.); Versus Arthritis UK Program (grant no. 21802, to A.R.C., M.K.-S. and A.E.); Versus Arthritis UK (grant no. 22272, to M.K.-S.); Wellcome Trust (no. 204820/Z/16/Z, to T.D.O. and M.K.-S.); and BPF_Medical Research Trust (to C.M.).
Author information
Authors and Affiliations
Contributions
S.A. and M.K.-S. conceived and oversaw the project, performed scRNA-seq, interpreted all the results and wrote the manuscript with feedback from all authors. S.A. and B.T. performed all synovial biopsies, ran synovial tissue processing/handling and linked clinical information with experimental datasets. L.M. analyzed all scRNA-seq and bulk RNA-seq data, validated scRNA-seq data by flow cytometry, together with M.K.-S., performed STM–FLS cocultures with the scRNA-seq BD Rhapsody system and helped write the manuscript. S.F. performed phenotyping of STMs, stimulation of STMs and the cocultures of MoM with FLS in the trans-well system. A.E. performed phenotyping of STMs, validation of scRNA-seq data with IHC/IF and direct coculture of MoM with FLS. S.A., E.G., M.R.G. and L.P. enrolled the study cohort. S.A., M.R.G., L.P., S.P. and A.L.F. followed all study cohorts and collected clinical and ultrasound data. S.A., M.R.G. and S.P. followed all patients in RA remission for withdrawal study in the SYNGem cohort. C.D.M. and S.P. performed serological assessment for autoantibody positivity of the entire study cohort. L.B. performed MerTK IHC staining of synovial biopsies. M.G. performed semiquantitative assessment of synovitis score in synovial biopsies. M.A. performed 10x Genomics of the Discovery Cohort. S.C. prepared the libraries of MoM–FLS cocultures. D.S. helped A.E. with validation of scRNA-seq classification of STMs by IF. S.N.S. provided bioinformatic input to scRNA-seq data. A.F. provided protocol for MoM–FLS coculture studies. C.M. helped with interpretation of data and writing of the manuscript. N.L.M. provided healthy synovial tissues. K.K. provided protocol for scRNA-seq. A.N., M.J.L. and C.P. provided data from the PEAC cohort. C.D.B., A.R.C., G.F., E.G. and I.B.M. helped with data interpretation. I.U. provided sorting protocol for scRNA-seq and helped with data interpretation. E.G. and I.B.M. assisted with running of the project. T.D.O. and M.K.-S. supervised all computational analysis in the study. All authors approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
All authors declare no competing interests.
Additional information
Peer review information Saheli Sadanand was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 A low proportion of MerTKposCD206pos STMs in remission was associated with increased risk of disease flare after treatment cessation.
a–d, Comparison of STM distribution between RA patients with disease remission defined by either DAS28 (n = 24) or Boolean criteria (n = 11). Analyses include comparison of STMs: a, single-marker positive or negative for CD206 or MerTK or CD163, b, double-marker positive or negative for MerTK and CD206, and double-marker positive or negative for CD206 and CD163, c, double-marker positive or negative for MerTK and CD163 and (d) triple-marker positive for MerTK, CD206 and CD163. e-i, Comparison of baseline STM distribution between RA patients in remission who subsequently flared (n = 11) or remained in remission (n = 11) after treatment discontinuation. Analyses include comparison of STMs: e, single-marker positive or negative for CD206 or MerTK or CD163, f, double-marker positive or negative for either MerTK or CD206, g, double-marker positive or negative for either CD163 or CD206, h, double-marker positive or negative for either MerTK or CD206 and (i) triple-marker positive for MerTK, CD206 and CD163. j, ROC curves for optimal cut-off values of STM proportions of CD206pos, MerTKposCD206pos, MerTKnegCD206neg, CD163posCD206pos, CD163negCD206neg, and the MerTKposCD206pos to MerTKnegCD206neg ratio discriminating disease flare in RA in remission (n = 22) described in e-i. (Wilson/Brown method) k, Comparison of occurrence of flare stratified by the cut-off values for different STM populations (Wilcoxon test.) Data in (a-i) are mean + /-sem, differences in STM populations between remission states were evaluated by Two-tailed Mann-Whitney, p-values provided on graphs.
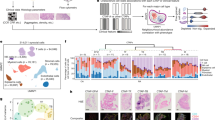
Extended Data Fig. 2 Integration of scRNAseq data.
a, Metadata for FACS sorted CD64posCD11bpos STMs sequenced in the Discovery Cohort (Cohort 1) at Oxford Genomics Centre. b, Metadata for all synovial cell types sequenced in the Validation Cohort (Cohort 2) at Glasgow Polyomics. c, Myeloid cells sequenced in Cohort 2 are separated computationally (based on positive expression of CD64, CD11b, CD14, MARCO, CD1c and LYZ), and integrated with synovial macrophages sequenced in the Discovery cohort 1.
Extended Data Fig. 3 Pathway analysis of differentially expressed genes between clusters revealing different effector pathways in STM subpopulations.
a, Heatmap illustrating scaled pseudo-bulk expression of significantly enriched pathways in four MerTKpos clusters and (b) in four MerTKneg clusters and in the MerTKpos ICAM1pos cluster (Healthy, n = 4; UPA, n = 4; naïve-active RA, n = 5; treatment-resistant RA, n = 6 and RA in remission, n = 6). Rows are genes and columns represent average expression for cells in each cluster by subject group. All genes are significantly expressed in at least 60% of cells in that cluster. DE Genes identified by Seurat function (MAST) were filtered afterwards to ensure that the p-value adjusted by Bonferroni correction is significant (p < 0.05). Average log fold change ≥ 0.25. Differentially expressed genes between clusters were used to perform GO and IPA analysis to identify significant cluster specific pathways (Fisher’s exact test with Bonferroni correction). Upregulated genes from selected significant pathways of interest are annotated.
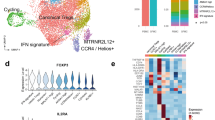
Extended Data Fig. 4 Comparison of Human and Mouse (Culemann et al., 2019) single-cell transcriptional profiling of synovial macrophages.
a, UMAP projections for human and mouse scRNAseq data analyzed separately. Mouse data clustered at a resolution of 0.3 and UMAP projection represents the top 12 PCs. Human data for this comparison included samples from healthy tissue (n = 4), UPA (n = 5) and naïve-active RA (n = 5) to align with disease conditions modelled in the mouse data (n = 4). b, Dendrogram representing the relationship between human macrophage phenotypes and mouse clusters. This plot was generated from the hierarchical clustering of the average expression of orthologous genes by each population. c-d, Patients’ cohort described in Fig. 2a. Violin plots show log-normalized expression values of STMN, a marker of proliferation; CSF1R, that is the highest in FOLR2posID2pos, and GM-CSFR (CSF2R) that is higher in MerTK negative clusters of human STMs. Shape of the violin represents the density of the data at different expression values with median marked by dot while colour represents unique STM cluster. d, MerTKpos STMs are enriched in tight-junction proteins. Heatmap illustrating scaled pseudobulk expression of significantly enriched pathways by each patient group within each of identified STM clusters. Rows represent genes with a potential contribution to synovial lining-layer barrier function (GO pathway- involved tight-junction assembly and organization). Columns represent equal average expression for cells in each cluster by subject group. The blue box highlights gene orthologue identified in mouse synovial lining macrophages as tight-junction proteins (Culemann et al). Among them, TJP1 is expressed by human MerTKposTREM2pos and MerTKposFOLR2posLYVE1pos STMs.
Extended Data Fig. 5 Flow-cytometric validation of STM clustering categorized by scRNAseq.
a–c, scRNAseq data (a) (patient cohort described in Fig. 2a) and flow cytometry data (b-c) representing 16 independent experiments with synovial tissue samples from 31 RA patients and 10 Healthy showing that mRNA and protein expression of MerTK and FOLR2 coincide, suggesting that FOLR2 can be used as an alternative marker of MerTKpos STMs. d, Representative gating strategies for TREM2 and LYVE1 positive STMs in conjunction with MerTK expression in health, active RA and RA in disease remission. The TREM2pos cluster is defined by the positive expression of MerTK and TREM2, and the LYVE1pos cluster is defined by the positive expression of LYVE1 and MerTK. A proportion of TREM2pos STMs are also LYVE1pos. The n numbers per staining and quantitative data are provided in Fig. 2i-k. e-g, scRNAseq data (e) (patient cohort described in Fig. 2a) and representative gating strategy for MerTKneg STMs (f) showing that most of MerTKneg STMs are CD48 positive. g, Distribution of CD9 and CLEC10a positive cells within MerTKnegCD48low/pos STMs in health, active RA and RA in disease remission are shown. The MerTKnegS100A12pos cluster is defined as CD48low/posCD9negCLEC10aneg; the MerTKnegSPP1pos cluster is defined as CD48pos/lowCD9posCLEC10aneg, and the MerTKnegCLEC10apos cluster is defined as CD48low/posCD9posCLEC10apos. The n numbers per staining and quantitative data are provided in Fig. 2i-k.
Extended Data Fig. 6 High-dimensional characterization of STMs using viSNE dimensionality reduction algorithm.
a,b, STMs in healthy and RA in sustained disease remission show enrichment in MerTK positive clusters (TREM2pos and FOLR2highLYVE1pos) while patients with active RA show an increase in MerTK negative clusters (CLEC10Apos, SPP1pos and S100A12pos clusters). a-b, Single cell synovial tissue digests from healthy controls and RA patients as described in Fig. 2i-k and Extended Data Fig. 5 were stained with panel 1 of 9 antibodies (plus dump panel) to identify MerTK positive clusters (a) or with panel 2 of 9 antibodies (plus dump panel) to identify MerTK negative clusters (Supplementary Table 8 and Methods) (b). viSNE plots of clustered total STMs (CD64posCD11bposlineageneg) are displayed for MerTK positive (a) and MerTK negative (b) STMs, showing cell density of clusters and changes between conditions. The number of cells per condition were normalized to 25 K. Bars represent individual expression scale for each marker. Dotted lines demarcate clusters dominant in remission RA/healthy (a) or active RA (b). a, Synovial tissue from Healthy (n = 9), active RA (n = 17) and RA in Remission (n = 13) were used to evaluate MerTKposFOLR2pos STMs; from healthy (n = 9), active RA (n = 17) and RA in Remission (n = 12) to evaluate MerTKposTREM2pos STMs; and Healthy (n = 9), active RA (n = 14) and RA in Remission (n = 9) to evaluate MerTKposLYVE2pos STMs. b, Synovial tissue from Healthy (n = 8), active RA (n = 13) and RA in Remission (n = 7) were used to evaluate MerTKnegCD48pos, MerTKnegS100A12pos and MerTKnegCLEC10apos STMs; and from Healthy (n = 7), active RA (n = 12) and RA in Remission (n = 7) to evaluate MerTKnegSPP1pos STMs.
Extended Data Fig. 7 TREM2pos STMs form a lining-layer in the healthy synovium and in the synovium from RA patients in sustained disease remission.
Representative confocal microscopy images (40×) showing IF staining for TREM2 (green) and macrophage marker CD68 (red) in (a) healthy synovium (b) active RA synovium, and (c) remission RA synovium. These show TREM2posCD68pos (solid white arrows) and TREM2negCD68pos (hollow white arrows) macrophages. The inset images show TREM2posCD68pos cells at higher magnification. Nuclei are stained with DAPI (blue). A, adipocyte. Images representative of synovial tissue of healthy donors (n = 5), active RA (n = 6) and remission RA (n = 6) obtained in 3 independent experiments with similar results are shown. Scale bars = 50 μm.
Extended Data Fig. 8 LYVE1pos STMs locate mainly in the synovial lining-layer in health and in remission RA, and predominantly in the interstitium in active RA.
Representative confocal microscopy images (40×) showing IF staining for LYVE1 (green) and macrophage marker CD68 (red) in (a) healthy synovium (b) active RA synovium (two different patients), and (c) remission RA synovium. These show LYVE1posCD68pos (solid white arrows) and LYVE1negCD68pos (hollow white arrows) macrophages. The inset images show LYVE1posCD68pos at higher magnification. Nuclei are stained with DAPI (blue). A, adipocyte; BV, blood vessel. Images representative of synovial tissue of healthy donors (n = 5), active RA (n = 6) and remission RA (n = 6) obtained in 3 independent experiments with similar results are shown. Scale bars = 50 μm.
Extended Data Fig. 9 CLEC10apos, S100A12pos and SPP1pos STMs are located predominantly in the synovial interstitium.
a–c, Representative confocal microscopy images (40×) showing IF staining for CLEC10a (green) and macrophage marker CD68 (red) in (a) healthy synovium (b) active RA synovium, and (c) remission RA synovium. These show CLEC10aposCD68pos (solid white arrows) and CLEC10anegCD68pos (hollow white arrows) macrophages, and CLEC10aposCD68neg cells that are not macrophages. Inset images show CLEC10aposCD68pos macrophages at higher magnification. d-f, Synovial S10012Apos STMs are scarce in healthy and remission RA but abundant in the sublining layer in active RA. Representative confocal microscopy images (40×) showing IHC staining for S10012A (green) and macrophage marker CD68 (red) in (d) healthy synovium (e) active RA synovium, and (f) remission RA synovium. These show S10012AposCD68pos (solid white arrows) macrophages only in active RA. S100A12AnegCD68pos (hollow white arrows) macrophages and S100A12AposCD68neg cells are located throughout the synovial tissue. Inset images show S10012AposCD68pos macrophages at higher magnification. g-i, Synovial SPP1pos STMs are scarce in healthy and remission RA but abundant in the sublining layer in active RA. Representative confocal microscopy images (40×) showing IF staining for SPP1 (green) and macrophage marker CD68 (red) in (g) healthy synovium, (h) active RA synovium (two patients), and (i) remission RA. These show SPP1posCD68pos (solid white arrows) macrophages and SPP1negCD68pos (hollow white arrows) macrophages. Inset images show SPP1posCD68pos macrophages at higher magnification. The nuclei are stained with DAPI (blue). Images representative of synovial tissue of healthy donors (n = 5), active RA (n = 6) and remission RA (n = 6) obtained in 3 independent experiments with similar results are shown. A, adipocyte; BV, blood vessel. Scale bars = 50 μm.
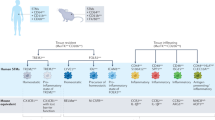
Extended Data Fig. 10 Distinct synovial tissue macrophage subsets regulate inflammation and remission in rheumatoid arthritis.
The HEALTHY synovial membrane (SM) contains predominantly MerTKpos STMs with two subpopulations: TREM2pos and LYVE1pos. Their transcriptomics suggest immunoregulatory functions, for example production of retinoic acid. In ACTIVE RA, the synovial membrane is infiltrated by MerTKnegCD48pos STMs with two main phenotypes, expressing either S100A alarmins and IL-1β, or osteopontin (SPP1); both are the main source of pathogenic TNF and IL-6, and potent contact-dependent inducers of chemokines and MMPs from synovial fibroblasts (FLS). RA in REMISSION is characterized by restoration of MerTKposTREM2pos and MerTKposLYVE1pos subpopulations. Their transcriptome is characterized by MerTK-dependent transcription factors that are negative-regulators of inflammation. They are low producers of pro-inflammatory cytokines; further downregulated by locally-produced GAS6. Instead they produce resolvins and induce a repair response in FLS. Their relative proportion in remission was indicative of flare after treatment cessation. When the proportion of MerTKpos STMs becomes less than 47.5%, (or the ratio of MerTKpos to MerTKneg becomes less than 2.5) there is a likelihood of FLARE after treatment cessation. MerTKneg STMs in patients predicted to flare have a CD48posS100A12pos phenotype that releases the alarmin S100A12 upon stimulation, suggesting a role in the initiation of flare. BM bone marrow; MMPs, matrix metalloproteinases; KLFs krueppel like factors; NR4As, nuclear receptor subfamily 4 group A; ATF3, cAMP-dependent transcription factor 3; TREM2, triggering receptor expressed on myeloid cells 2; LYVE1, lymphatic vessels endothelial hyaluronan receptor 1; FOLR2, folate receptor beta; GAS6, growth arrest-specific 6; S100A12, S100 calcium-binding protein A12; THY1, CD90.
Supplementary information
Supplementary Information
Supplementary Figs. 1–9 and Tables 1–11.
Supplementary Data 1
Metrics of scRNA-seq data and complete lists of DE genes for Figs. 2 and 4–6, Extended Data Fig. 3 and Supplementary Figs. 4 and 7.
Rights and permissions
About this article
Cite this article
Alivernini, S., MacDonald, L., Elmesmari, A. et al. Distinct synovial tissue macrophage subsets regulate inflammation and remission in rheumatoid arthritis. Nat Med 26, 1295–1306 (2020). https://doi.org/10.1038/s41591-020-0939-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-020-0939-8
This article is cited by
-
Profiling joint tissues at single-cell resolution: advances and insights
Nature Reviews Rheumatology (2024)
-
ATF3 coordinates the survival and proliferation of cardiac macrophages and protects against ischemia–reperfusion injury
Nature Cardiovascular Research (2024)
-
Joint-specific memory, resident memory T cells and the rolling window of opportunity in arthritis
Nature Reviews Rheumatology (2024)
-
Using explainable artificial intelligence to predict and forestall flare in rheumatoid arthritis
Nature Medicine (2024)
-
Axl and MerTK regulate synovial inflammation and are modulated by IL-6 inhibition in rheumatoid arthritis
Nature Communications (2024)