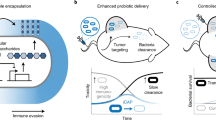
Abstract
Synthetic biology is driving a new era of medicine through the genetic programming of living cells1,2. This transformative approach allows for the creation of engineered systems that intelligently sense and respond to diverse environments, ultimately adding specificity and efficacy that extends beyond the capabilities of molecular-based therapeutics3,4,5,6. One particular area of focus has been the engineering of bacteria as therapeutic delivery systems to selectively release therapeutic payloads in vivo7,8,9,10,11. Here we engineered a non-pathogenic Escherichia coli strain to specifically lyse within the tumor microenvironment and release an encoded nanobody antagonist of CD47 (CD47nb)12, an anti-phagocytic receptor that is commonly overexpressed in several human cancer types13,14. We show that delivery of CD47nb by tumor-colonizing bacteria increases activation of tumor-infiltrating T cells, stimulates rapid tumor regression, prevents metastasis and leads to long-term survival in a syngeneic tumor model in mice. Moreover, we report that local injection of CD47nb-expressing bacteria stimulates systemic tumor-antigen-specific immune responses that reduce the growth of untreated tumors, providing proof-of-concept for an abscopal effect induced by an engineered bacterial immunotherapy. Thus, engineered bacteria may be used for safe and local delivery of immunotherapeutic payloads leading to systemic antitumor immunity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All requests for raw and analyzed data and materials will be promptly reviewed by Columbia Technology Ventures to verify whether the request is subject to any intellectual property or confidentiality obligations. Any data and materials that can be shared will be released through a material transfer agreement.
References
Fischbach, M. A., Bluestone, J. A. & Lim, W. A. Cell-based therapeutics: the next pillar of medicine. Sci. Transl. Med. 5, 179ps177 (2013).
Weber, W. & Fussenegger, M. Emerging biomedical applications of synthetic biology. Nat. Rev. Genet. 13, 21–35 (2012).
Lim, W. A. & June, C. H. The principles of engineering immune cells to treat cancer. cell 168, 724–740 (2017).
Ruder, W. C., Lu, T. & Collins, J. J. Synthetic biology moving into the clinic. Science 333, 1248–1252 (2011).
Chen, Y. Y. & Smolke, C. D. From DNA to targeted therapeutics: bringing synthetic biology to the clinic. Sci. Transl. Med. 3, 106ps142 (2011).
Wu, M.-R., Jusiak, B. & Lu, T. K. Engineering advanced cancer therapies with synthetic biology. Nat. Rev. Cancer 19, 187–195 (2019).
Chien, T., Doshi, A. & Danino, T. Advances in bacterial cancer therapies using synthetic biology. Curr. Opin. Syst. Biol. 5, 1–8 (2017).
Din, M. O. et al. Synchronized cycles of bacterial lysis for in vivo delivery. Nature 536, 81–85 (2016).
Pedrolli, D. B., Ribeiro, N. V., Squizato, P. N., de Jesus, V. N. & Cozetto, D. A. Engineering microbial living therapeutics: the synthetic biology toolbox. Trends Biotechnol. 37, 100–115 (2019).
Zhou, S., Gravekamp, C., Bermudes, D. & Liu, K. Tumour-targeting bacteria engineered to fight cancer. Nat. Rev. Cancer 18, 727–743 (2018).
Helmink, B. A., Khan, M. A. W., Hermann, A., Gopalakrishnan, V. & Wargo, J. A. The microbiome, cancer, and cancer therapy. Nat. Med. 25, 377–388 (2019).
Sockolosky, J. T. et al. Durable antitumor responses to CD47 blockade require adaptive immune stimulation. Proc. Natl Acad. Sci. USA 113, E2646–E2654 (2016).
Majeti, R. et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell 138, 286–299 (2009).
Willingham, S. B. et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc. Natl Acad. Sci. USA 109, 6662–6667 (2012).
Coley, W. B. Contribution to the knowledge of sarcoma. Ann. Surg. 14, 199–220 (1891).
Berendt, M. J., North, R. J. & Kirstein, D. P. The immunological basis of endotoxin-induced tumor regression. Requirement for T-cell-mediated immunity. J. Exp. Med. 148, 1550–1559 (1978).
Tsung, K. & Norton, J. A. Lessons from Coley’s toxin. Surg. Oncol. 15, 25–28 (2006).
Mellman, I., Coukos, G. & Dranoff, G. Cancer immunotherapy comes of age. Nature 480, 480–489 (2011).
Jiang, S. N. et al. Inhibition of tumor growth and metastasis by a combination of Escherichia coli-mediated cytolytic therapy and radiotherapy. Mol. Ther. 18, 635–642 (2010).
Malmgren, R. A. & Flanigan, C. C. Localization of the vegetative form of Clostridium tetani in mouse tumors following intravenous spore administration. Cancer Res. 15, 473–478 (1955).
Brown, J. M. & Wilson, W. R. Exploiting tumour hypoxia in cancer treatment. Nat. Rev. Cancer 4, 437–447 (2004).
Forbes, N. S. Engineering the perfect (bacterial) cancer therapy. Nat. Rev. Cancer 10, 785–794 (2010).
Zheng, J. H. et al. Two-step enhanced cancer immunotherapy with engineered Salmonella typhimurium secreting heterologous flagellin. Sci. Transl. Med. 9, eaak9537 (2017).
Gardner, T. S., Cantor, C. R. & Collins, J. J. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000).
Basu, S., Gerchman, Y., Collins, C. H., Arnold, F. H. & Weiss, R. A synthetic multicellular system for programmed pattern formation. Nature 434, 1130–1134 (2005).
Friedland, A. E. et al. Synthetic gene networks that count. Science 324, 1199–1202 (2009).
Danino, T., Mondragon-Palomino, O., Tsimring, L. & Hasty, J. A synchronized quorum of genetic clocks. Nature 463, 326–330 (2010).
Elowitz, M. B. & Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000).
Jaiswal, S. et al. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell 138, 271–285 (2009).
Liu, X. et al. CD47 blockade triggers T cell-mediated destruction of immunogenic tumors. Nat. Med. 21, 1209–1215 (2015).
Kauder, S. E. et al. ALX148 blocks CD47 and enhances innate and adaptive antitumor immunity with a favorable safety profile. PLoS ONE 13, e0201832 (2018).
Liu, X. et al. Dual targeting of innate and adaptive checkpoints on tumor cells limits immune evasion. Cell Rep. 24, 2101–2111 (2018).
Ingram, J. R. et al. Localized CD47 blockade enhances immunotherapy for murine melanoma. Proc. Natl Acad. Sci. USA 114, 10184–10189 (2017).
Huang, Y., Ma, Y., Gao, P. & Yao, Z. Targeting CD47: the achievements and concerns of current studies on cancer immunotherapy. J. Thorac. Dis. 9, E168–E174 (2017).
Advani, R. et al. CD47 blockade by Hu5F9-G4 and rituximab in non-Hodgkin’s lymphoma. N. Engl. J. Med. 379, 1711–1721 (2018).
Veillette, A. & Chen, J. SIRPα–CD47 immune checkpoint blockade in anticancer therapy. Trends Immunol. 39, 173–184 (2018).
Skinner, S. O., Sepulveda, L. A., Xu, H. & Golding, I. Measuring mRNA copy number in individual Escherichia coli cells using single-molecule fluorescent in situ hybridization. Nat. Protoc. 8, 1100–1113 (2013).
Kong, X. N. et al. LPS-induced down-regulation of signal regulatory protein α contributes to innate immune activation in macrophages. J. Exp. Med. 204, 2719–2731 (2007).
Danino, T. et al. Programmable probiotics for detection of cancer in urine. Sci. Transl. Med. 7, 289ra84 (2015).
Armstrong, A. C. et al. Immunization with a recombinant adenovirus encoding a lymphoma idiotype: induction of tumor-protective immunity and identification of an idiotype-specific T cell epitope. J. Immunol. 168, 3983–3991 (2002).
Haldimann, A. & Wanner, B. L. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183, 6384–6393 (2001).
Gerdes, K. et al. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 5, 2023–2029 (1986).
Derman, A. I. et al. Alp7R regulates expression of the actin-like protein Alp7A in Bacillus subtilis. J. Bacteriol. 194, 2715–2724 (2012).
Acknowledgements
This work was supported by the NIH Pathway to Independence Award (R00CA197649–02) (to T.D.), DoD Idea Development Award (LC160314) (to T.D.), DoD Era of Hope Scholar Award (BC160541) (to T.D.), NIH R01GM069811 (to T.D.), NIH K22AI127847 (to N.A.), Searle Scholars Program SSP-2017-2179 (to N.A.), Bonnie J. Addario Lung Cancer Foundation Young Investigators Team Award (to N.A. and T.D.) and the Roy and Diana Vagelos Precision Medicine Pilot Grant (to N.A. and T.D.). Research reported in this publication was performed in the Columbia University Department of Microbiology and Immunology Flow Cytometry Core facility. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank K. T. Fortson and O. Velasquez for technical assistance with flow cytometry experiments and in vivo tumor experiments, respectively; M. O. Din for input pertaining to SLC characterization experiments; and V. Yeong and members of the Obermeyer group for assistance with affinity chromatography and protein purification. We thank R. L. Vincent, T. M. Savage, K. A. Kaiser and L. F. Loffredo for critical review of the manuscript.
Author information
Authors and Affiliations
Contributions
S. Chowdhury, N.A. and T.D. conceived and designed the study. S. Chowdhury, S. Castro, C.C. and T.E.H. performed in vivo experiments. S. Chowdhury performed in vitro characterization of eSLC–CD47nb and conducted immunophenotyping experiments. S. Chowdhury, N.A. and T.D. analyzed data and wrote the manuscript with input from all other authors.
Corresponding authors
Ethics declarations
Competing interests
S. Chowdhury, N.A. and T.D. have filed a provisional patent application with the US Patent and Trademark Office (US Patent Application No. 62/747,826) related to this work. T.D. and N.A. have a financial interest in GenCirq, Inc.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peer review information: Saheli Sadanand and Joao Monteiro were the primary editors on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Extended data
Extended Data Fig. 1 Map of plasmids used in this study.
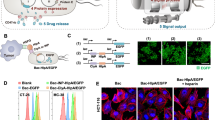
a, pSC01, single plasmid with synchronized lysis circuit. b, pSC02, stabilized plasmid that drives constitutive expression of HA-tagged anti-CD47 nanobody. c, pSC03, empty vector control.
Extended Data Fig. 2 E. coli that are capable of synchronized lysis produce functional anti-CD47 nanobodies.
a, Bacterial growth dynamics over time in agar-pad microscopy experiments. b, Immunoblot of bacterial culture supernatants (S) and cell pellets (P) in strains with and without SLC designed to constitutively produce the HA-tagged CD47 nanobody. a.u., arbitrary units. Data are representative of three independent experimental replicates. The immunoblot has been cropped to show relevant bands; a molecular weight marker is indicated on the right (in kDa). The uncropped blot is available as Source Data. c, A20 cells were co-incubated with a fixed concentration of FITC-conjugated isotype control (IgG2a–FITC) antibody along with increasing concentrations of bacterial lysates of bacteria that constitutively expressed CD47nb or empty vector. Data are representative of two independent experimental replicates. d, A20 cells were co-incubated with a fixed concentration of FITC-conjugated anti-CD47 (miap301) antibody along with serial dilutions of recombinant 6×His-tagged rCD47nb. Data are representative of two independent experimental replicates. e, In vitro phagocytosis by BMDMs of DiI-labeled A20 cells pretreated with PBS, miap301 or IgG2a isotype control, or with serial dilutions of eSLC or eSLC–CD47nb lysates in PBS. Data are mean ± s.e.m., n = 4 fields of view from three technical replicates.
Extended Data Fig. 3 Individual kinetics of intratumoral bacterial immunotherapy.
a, Individual tumor growth trajectories (n = 7 mice per group); related to data shown in Fig. 2a. b, Representative images of subcutaneous A20-tumor-bearing BALB/c mice treated with PBS, eSLC or eSLC–CD47nb. c, Relative body weight of A20-tumor-bearing BALB/c mice over time (n = 7 mice per group; P > 0.05, two-way ANOVA with Tukey’s multiple comparisons test). Data are mean ± s.e.m. d, Individual tumor growth trajectories after treatment with eSLC (intratumoral injection (IT)), miap301 (intraperitoneal injection (IP)), eSLC (intratumoral injection) and miap301 (intraperitoneal injection), eSLC–CD47nb lysate, rCD47nb (intratumoral injection) or eSLC–CD47nb (intratumoral injection); related to data shown in Fig. 2e.
Extended Data Fig. 4 Immunotherapeutic bacteria limit tumor growth in syngeneic mouse models of melanoma and triple-negative breast cancer.
a, Relative body weight of B16-F10-bearing C57BL/6 mice over time. Mice were treated with PBS (n = 4 mice), eSLC (n = 5 mice), miap301 (n = 5 mice) or eSLC–CD47nb (n = 6 mice) (P < 0.05, two-way ANOVA with Tukey’s multiple comparisons test). Data are mean ± s.e.m. b, Individual tumor growth trajectories of subcutaneous 4T1 tumors after intratumoral injection of eSLC (n = 7 tumors) or eSLC–CD47nb (n = 8 tumors); related to data shown in Fig. 2f. c, Individual tumor growth trajectories of subcutaneous B16-F10 melanoma after intraperitoneal injection of miap301 or intratumoral injection of PBS, eSLC or eSLC–CD47nb; related to data shown in Fig. 2h.
Extended Data Fig. 5 Intravenous bacterial immunotherapy limits tumor growth in mice in a subcutaneous A20 lymphoma model.
a, Individual tumor growth trajectories of subcutaneous A20 tumors after intraperitoneal injection of miap301 (n = 10 tumors) or intravenous injection of eSLC (n = 9 tumors) or eSLC–CD47nb (n = 10 tumors); related to data shown in Fig. 2i. b, Biodistribution of eSLC–CD47nb E. coli on day 8 after the final intravenous bacterial treatment. Excised tumors, livers, spleens and kidneys were homogenized, serially diluted and plated onto LB–agar plates. Colonies were counted to determine the CFU g−1 of tissue (n = 3 per group). Data are mean ± s.e.m. c, Relative body weight of A20-tumor-bearing BALB/c mice that received intravenous injections of bacteria (eSLC, n = 5 mice; eSLC–CD47nb, n = 4 mice) or intraperitoneal injections of miap301 (n = 5 mice) (P > 0.05, two-way ANOVA with Tukey’s multiple comparisons test). Data are mean ± s.e.m.
Extended Data Fig. 6 Immunophenotyping of tumor-infiltrating myeloid and lymphoid cell subsets after intratumoral injection of bacteria.
First, 5 × 106 A20 cells were subcutaneously implanted into the hind flanks of BALB/c mice. Subsequently, when tumors reached 100–150 mm3 in volume (day 0), mice were treated with PBS, eSLC or eSLC–CD47nb on days 0, 4 and 7. On day 3 or day 8, tumors were homogenized and tumor-infiltrating myeloid and lymphoid cell subsets were isolated for flow cytometry analysis. a, Frequency of isolated MHC class IIhiCD11b+F4/80+ macrophages on day 3 after treatment (n = 5 tumors per group, *P = 0.0271, unpaired two-tailed t-test). b, Mean fluorescence intensity (MFI) of SIRPα staining within the CD11b+F4/80+ subset on day 8 after treatment (n = 5 tumors per group, *P = 0.0209, unpaired two-tailed t-test). c,d, Frequencies of CTLA4+ cells within the FOXP3−CD4+ T cell population (c; n = 5 tumors per group, **P = 0.0018, unpaired two-tailed t-test) and CD8+ T cell population (d; n = 5 tumors per group, **P = 0.0098, unpaired two-tailed t-test). e,f, Frequencies of TNF+ cells within FOXP3−CD4+ T cell (e) and CD8+ T cell (f) populations following ex vivo stimulation (ns, not significant (P > 0.05), n = 3 tumors per group, unpaired two-tailed t-test). g, Frequency of IL-17+ cells within the FOXP3−CD4+ T cell population following ex vivo stimulation (*P = 0.0402, n = 3 tumors per group, unpaired two-tailed t-test). h, Frequency of IFNγ+ cells within the CD8+ T cell population following ex vivo stimulation (n = 3 tumors per group, P > 0.05, unpaired two-tailed t-test). All data are mean ± s.e.m.
Extended Data Fig. 7 Immunotherapeutic bacteria lead to increased IFNγ production by splenic T cells after stimulation with tumor antigens.
Enzyme-linked immunosorbent assay (ELISA) of IFNγ using supernatants from overnight co-incubation of splenocytes isolated from each of the indicated treatment groups with irradiated A20 cells (n = 2 mice per group of three technical replicates).
Extended Data Fig. 8 Intratumoral bacterial immunotherapy leads to distal tumor control.
a, Individual tumor growth trajectories of treated (injected) and untreated A20 tumors after intratumoral injection of PBS, eSLC or eSLC–CD47nb; related to Fig. 4a,b. b, SLC− and SLC+ E. coli NisLux were intratumorally injected into a single flank of A20-tumor-bearing mice (scale represents radiance (p s−1 cm−2 sr−1)). Luminescence was measured over time using in vivo imaging. Representative image of mouse no. 3 from each group over time. c, Luminescence heat maps over time (n = 5 mice per group). Colors represent average radiance (p s−1 cm−2 sr−1).
Extended Data Fig. 9 Immunophenotyping of tumor-infiltrating lymphocytes in untreated tumors after single-flank injection of bacteria.
First, 5 × 106 A20 cells were implanted into the hind flanks of BALB/c mice. Subsequently, when tumors reached ~100 mm3 in volume (day 0), mice were treated with PBS, eSLC or eSLC–CD47nb on day 0, 4 and 7 by injection into a single tumor. Untreated tumors were extracted and analyzed by flow cytometry on day 8. n = 5 mice per group. a, Frequency of Ki-67+ cells within the FOXP3−CD4+ T cell population (P > 0.05, unpaired two-tailed t-test). b,c, Frequency of tumor-infiltrating IFNγ+ cells within the FOXP3−CD4+ T cell population (b; *P = 0.0327, unpaired two-tailed t-test) and CD8+ T cell population (c; *P = 0.0104, unpaired two-tailed t-test) following ex vivo stimulation with PMA and ionomycin in the presence of brefeldin A. d,e, Frequencies of CTLA4+ cells within the FOXP3−CD4+ T cell population (d; P > 0.05, unpaired two-tailed t-test) and CD8+ T cell population (e; *P = 0.0259, unpaired two-tailed t-test). f, Frequency of tumor-infiltrating IFNγ+ cells within the FOXP3−CD4+ T cell population following ex vivo restimulation with A20-Id peptide (DYWGQGTEL) in the presence of brefeldin A (P > 0.05, unpaired two-tailed t-test). All data are mean ± s.e.m.
Extended Data Fig. 10 Distal tumor control requires SLC+ bacteria engineered to produce CD47nb.
a,b, Individual tumor growth trajectories of treated (injected) and untreated A20 tumors after intratumoral injection of eSLC, eCD47nb or eSLC–CD47nb (n = 4 mice per group); related to Fig. 4h,i.
Supplementary information
Source data
Source Data Extended Data Fig. 2b
Unprocessed Western Blot for Extended Data Fig. 2b.
Rights and permissions
About this article
Cite this article
Chowdhury, S., Castro, S., Coker, C. et al. Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat Med 25, 1057–1063 (2019). https://doi.org/10.1038/s41591-019-0498-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-019-0498-z
This article is cited by
-
Cas9-assisted biological containment of a genetically engineered human commensal bacterium and genetic elements
Nature Communications (2024)
-
Promising dawn in tumor microenvironment therapy: engineering oral bacteria
International Journal of Oral Science (2024)
-
Live bacterial therapeutics for detection and treatment of colorectal cancer
Nature Reviews Gastroenterology & Hepatology (2024)
-
Roles of exosomes in immunotherapy for solid cancers
Cell Death & Disease (2024)
-
Camouflaging attenuated Salmonella by cryo-shocked macrophages for tumor-targeted therapy
Signal Transduction and Targeted Therapy (2024)