Abstract
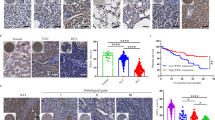
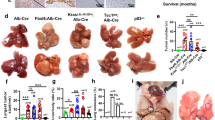
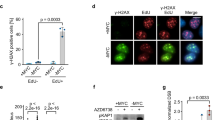
Cancer cells develop mechanisms to escape immunosurveillance, among which modulating the expression of immune suppressive messenger RNAs is most well-documented. However, how this is molecularly achieved remains largely unresolved. Here, we develop an in vivo mouse model of liver cancer to study oncogene cooperation in immunosurveillance. We show that MYC overexpression (MYCTg) synergizes with KRASG12D to induce an aggressive liver tumor leading to metastasis formation and reduced mouse survival compared with KRASG12D alone. Genome-wide ribosomal footprinting of MYCTg;KRASG12 tumors compared with KRASG12D revealed potential alterations in translation of mRNAs, including programmed-death-ligand 1 (PD-L1). Further analysis revealed that PD-L1 translation is repressed in KRASG12D tumors by functional, non-canonical upstream open reading frames in its 5′ untranslated region, which is bypassed in MYCTg;KRASG12D tumors to evade immune attack. We show that this mechanism of PD-L1 translational upregulation was effectively targeted by a potent, clinical compound that inhibits eIF4E phosphorylation, eFT508, which reverses the aggressive and metastatic characteristics of MYCTg;KRASG12D tumors. Together, these studies reveal how immune-checkpoint proteins are manipulated by distinct oncogenes at the level of mRNA translation, which can be exploited for new immunotherapies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The sequencing data of the manuscript can be accessed using the following accession number: GSE105147.
References
Chen, D. S. & Mellman, I. Oncology meets immunology: the cancer-immunity cycle. Immunity 39, 1–10 (2013).
Casey, S. C. et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 352, 227–231 (2016).
Marzec, M. et al. Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD-L1, B7-H1). Proc. Natl Acad. Sci. USA 105, 20852–20857 (2008).
Stewart, B. W. & Wild, C. World Cancer Report 2014 (International Agency for Research on Cancer WHO Press, 2014).
Ally, A. et al. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 169, 1327 (2017).
Schlaeger, C. et al. Etiology-dependent molecular mechanisms in human hepatocarcinogenesis. Hepatology 47, 511–520 (2008).
Marquardt, J. U. et al. Sequential transcriptome analysis of human liver cancer indicates late stage acquisition of malignant traits. J. Hepatol. 60, 346–353 (2014).
Kaposi-Novak, P. et al. Central role of c-Myc during malignant conversion in human hepatocarcinogenesis. Cancer Res. 69, 2775–2782 (2009).
O’Dell, M. R. et al. Kras(G12D) and p53 mutation cause primary intrahepatic cholangiocarcinoma. Cancer Res. 72, 1557–1567 (2012).
Saha, S. K. et al. Mutant IDH inhibits HNF-4alpha to block hepatocyte differentiation and promote biliary cancer. Nature 513, 110–114 (2014).
Grivennikov, S. I., Greten, F. R. & Karin, M. Immunity, inflammation, and cancer. Cell 140, 883–899 (2010).
Sato, E. et al. Intraepithelial CD8+tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc. Natl Acad. Sci. USA 102, 18538–18543 (2005).
Gao, Q. et al. Intratumoral balance of regulatory and cytotoxic T cells is associated with prognosis of hepatocellular carcinoma after resection. J. Clin. Oncol. 25, 2586–2593 (2007).
Fu, J. L. et al. Increased regulatory T cells correlate with CD8 T-cell impairment and poor survival in hepatocellular carcinoma patients. Gastroenterology 132, 2328–2339 (2007).
Fridman, W. H., Pages, F., Sautes-Fridman, C. & Galon, J. The immune contexture in human tumours: impact on clinical outcome. Nat. Rev. Cancer 12, 298–306 (2012).
Tumeh, P. C. et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014).
Mellman, I., Coukos, G. & Dranoff, G. Cancer immunotherapy comes of age. Nature 480, 480–489 (2011).
Joyce, J. A. & Fearon, D. T. T cell exclusion, immune privilege, and the tumor microenvironment. Science 348, 74–80 (2015).
Kortlever, R. M. et al. Myc cooperates with Ras by programming inflammation and immune suppression. Cell 171, 1301–1315 e1314 (2017).
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. S. & Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009).
Butte, M. J., Keir, M. E., Phamduy, T. B., Sharpe, A. H. & Freeman, G. J. Programmed death-1 ligand 1 interacts specifically with the B7-1 costimulatory molecule to inhibit T cell responses. Immunity 27, 111–122 (2007).
Herbst, R. S. et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 515, 563–567 (2014).
Gordon, S. R. et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature 545, 495–499 (2017).
Shalapour, S. et al. Immunosuppressive plasma cells impede T-cell-dependent immunogenic chemotherapy. Nature 521, 94–U235 (2015).
Khan, A. R. et al. PD-L1hi B cells are critical regulators of humoral immunity. Nat. Commun. 6, 5997 (2015).
Lau, J. et al. Tumour and host cell PD-L1 is required to mediate suppression of anti-tumour immunity in mice. Nat. Commun. 8, 14572 (2017).
Gollwitzer, E. S. et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat. Med. 20, 642–647 (2014).
Truitt, M. L. & Ruggero, D. New frontiers in translational control of the cancer genome. Nat. Rev. Cancer 17, 332 (2017).
Sendoel, A. et al. Translation from unconventional 5′ start sites drives tumour initiation. Nature 541, 494–499 (2017).
Stern-Ginossar, N. et al. Decoding human cytomegalovirus. Science 338, 1088–1093 (2012).
Calkhoven, C. F., Muller, C. & Leutz, A. Translational control of C/EBP alpha and C/EBP beta isoform expression. Gene Dev. 14, 1920–1932 (2000).
Palam, L. R., Baird, T. D. & Wek, R. C. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J. Biol. Chem. 286, 10939–10949 (2011).
Starck, S. R. et al. Translation from the 5′ untranslated region shapes the integrated stress response.Science 351, 3867 (2016).
Sekine, Y. et al. Stress responses. Mutations in a translation initiation factor identify the target of a memory-enhancing compound. Science 348, 1027–1030 (2015).
Sidrauski, C., McGeachy A. M. & Ingolia, N. T. & Walter, P. The small molecule ISRIB reverses the effects of eIF2alpha phosphorylation on translation and stress granule assembly. Elife 4, 05033 (2015).
Furic, L. et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc. Natl Acad. Sci. USA 107, 14134–14139 (2010).
Herdy, B. et al. Translational control of the activation of transcription factor NF-kappaB and production of type I interferon by phosphorylation of the translation factor eIF4E. Nat. Immunol. 13, 543–550 (2012).
Ueda, T., Watanabe-Fukunaga, R., Fukuyama, H., Nagata, S. & Fukunaga, R. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol. Cell. Biol. 24, 6539–6549 (2004).
Reich, S. H. et al. Structure-based design of pyridone-aminal eFT508 targeting dysregulated translation by selective mitogen-activated protein kinase interacting kinases 1 and 2 (MNK1/2) inhibition. J. Med. Chem. 26, 3516–3540 (2018).
Pulko, V. et al. B7-H1 expressed by activated CD8 T cells is essential for their survival. J. Immunol. 187, 5606–5614 (2011).
Liu, X. et al. B7-H1 antibodies lose antitumor activity due to activation of p38 MAPK that leads to apoptosis of tumor-reactive CD8(+) T cells. Sci. Rep. 6, 36722 (2016).
Betts, M. R. et al. Sensitive and viable identification of antigen-specific CD8+T cells by a flow cytometric assay for degranulation. J. Immunol. Methods 281, 65–78 (2003).
Hart, L. S. et al. ER stress-mediated autophagy promotes Myc-dependent transformation and tumor growth. J. Clin. Invest. 122, 4621–4634 (2012).
Kataoka, K. et al. Aberrant PD-L1 expression through 3′-UTR disruption in multiple cancers. Nature 534, 402 (2016).
Cerezo, M. et al. Translational control of tumor immune escape via the eIF4F-STAT1-PD-L1 axis in melanoma. Nat. Med. 24, 1877–1886 (2018).
D’Cruz, C. M. et al. c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nat. Med. 7, 235–239 (2001).
Ying, H. Q. et al. Oncogenic kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670 (2012).
Sharma, P., Hu-Lieskovan, S., Wargo, J. A. & Ribas, A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168, 707–723 (2017).
Matsuda, T. & Cepko, C. L. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc. Natl Acad. Sci. USA 101, 16–22 (2004).
Miyazaki, J. et al. Expression vector system based on the chicken beta-actin promoter directs efficient production of interleukin-5. Gene 79, 269–277 (1989).
Jackson, E. L. et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Gene Dev. 15, 3243–3248 (2001).
Rees, S. et al. Bicistronic vector for the creation of stable mammalian cell lines that predisposes all antibiotic-resistant cells to express recombinant protein. Biotechniques 20, 102–104, 106, 108–110 (1996).
Okada, A., Lansford, R., Weimann, J. M., Fraser, S. E. & McConnell, S. E. Imaging cells in the developing nervous system with retrovirus expressing modified green fluorescent protein. Exp. Neurol. 156, 394–406 (1999).
Hsieh, A. C. et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485, 55–61 (2012).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–U354 (2012).
Chen, Y., Lun, A. T. & Smyth, G. K. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res. 5, 1438 (2016).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Law, C. W., Chen, Y. S., Shi, W. & Smyth, G. K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014).
Robinson, M. D. & Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Bindea, G. et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093 (2009).
Szklarczyk, D. et al. STRINGv10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 (2015).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Liu, F., Song, Y. & Liu, D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 6, 1258–1266 (1999).
Kozak, M. Primer extension analysis of eukaryotic ribosome-mRNA complexes. Nucleic Acids Res. 26, 4853–4859 (1998).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Acknowledgements
We would like to thank members of the Ruggero laboratory for discussion and critical reading of the manuscript. We thank C. Her for his help with the mouse HCC cell line generation protocol, D. Wang for the help with hydrodynamic transfection, and K. Fujii and D. Simsek for advice and technique assistance. We thank B. Tiano for the contribution in developing and characterizing the MYC transgenic mice. This work was supported by the Damon Runyon Postdoctoral Fellowship (Y.X.), AACR-Incyte Corporation Fellowship In Basic Cancer Research (grant no. 17-40-46-JIN) (H.Y.J), Life Science Foundation Postdoctoral Fellowship (S.Z.), Department of Defense Physician training award (H.G.N), the Campini Foundation, The Leukemia and Lymphoma Foundation Career Development Grant and UCSF Department of Pediatrics K12 (grant no. 5K12HDO72222-05) (C.M.F.), American Cancer Society Postdoctoral Fellowship and Conquer Cancer Foundation Young Investigator Award (J.D.G), Pew Scholars Award (M.B.), NIH grant no. 1R01HD086634 (M.B.), NIH grants (nos. R01CA140456, R01CA154916, and R01CA184624) (D.R.). M.B. is a New York Stem Cell Foundation Robertson Investigator. D.R. is a Leukemia and Lymphoma Society Scholar.
Author information
Authors and Affiliations
Contributions
D.R. conceived and supervised the project. Y.X. and M.P. designed and performed most experiments with the help of the other authors. H.Y.J. contributed to the immune profiling analyses, luciferase reporter assays, hydrodynamic tail vein injection and flow cytometry analyses. C.M.F. performed the immune profiling. Y.W., E.D., and A.G. contributed to western blots, cloning, qPCR, CRISPR cell line generation, and intrahepatic HCC graft implantation. J.T.C. developed the MYC transgenic mice and helped C.R.S. with the ribosome profiling and RNA sequencing. L.S. helped with the ribosome-profiling sample preparation. S.A. helped with genome alignment. M.B. and Z.S. performed the bioinformatics analyses related to ribosome profiling and RNA-Seq. H.G.N. and L.X. performed immunofluorescence staining. H.G.N., S.E.U., and J.D.G. provided pathology support and provided human HCC primary samples. K.R.W., S.H.R., and S.T.W. developed and/or supported the development of eFT508. Y.X. and D.R. wrote the manuscript with contributions from M.B., H.Y.J., and C.M.F.
Corresponding author
Ethics declarations
Competing interests
C.R.S., S.H.R. and K.R.W. are employees and shareholders of eFFECTOR Therapeutics, Inc. S.T.W. is the President and CEO of eFFECTOR Therapeutics, Inc. D.R. is a shareholder of eFFECTOR Therapeutics, Inc., and a member of its scientific advisory board.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15
Supplementary Table 1
The transcriptional and translational profiles of gene expression in wild-type (WT) and KRAS-G12D liver tumors
Supplementary Table 2
The transcriptional and translational profiles of gene expression in KRAS-G12D and MYC-Tg;KRAS-G12D liver tumors
Supplementary Table 3
Enriched gene ontology categories (biological processes) among 339 transcripts that are translationally induced comparing MYCTg;KRAS-G12D to KRAS-G12D
Rights and permissions
About this article
Cite this article
Xu, Y., Poggio, M., Jin, H.Y. et al. Translation control of the immune checkpoint in cancer and its therapeutic targeting. Nat Med 25, 301–311 (2019). https://doi.org/10.1038/s41591-018-0321-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-018-0321-2
This article is cited by
-
Translating tumour immunity control
Nature Reviews Cancer (2024)
-
A lncRNA Dleu2-encoded peptide relieves autoimmunity by facilitating Smad3-mediated Treg induction
EMBO Reports (2024)
-
Adjuvant and neoadjuvant immunotherapies in hepatocellular carcinoma
Nature Reviews Clinical Oncology (2024)
-
Elevated FBXL6 activates both wild-type KRAS and mutant KRASG12D and drives HCC tumorigenesis via the ERK/mTOR/PRELID2/ROS axis in mice
Military Medical Research (2023)
-
MYC in liver cancer: mechanisms and targeted therapy opportunities
Oncogene (2023)