Abstract
Two microglial TAM receptor tyrosine kinases, Axl and Mer, have been linked to Alzheimer’s disease, but their roles in disease have not been tested experimentally. We find that in Alzheimer’s disease and its mouse models, induced expression of Axl and Mer in amyloid plaque–associated microglia was coupled to induced plaque decoration by the TAM ligand Gas6 and its co-ligand phosphatidylserine. In the APP/PS1 mouse model of Alzheimer’s disease, genetic ablation of Axl and Mer resulted in microglia that were unable to normally detect, respond to, organize or phagocytose amyloid-β plaques. These major deficits notwithstanding, TAM-deficient APP/PS1 mice developed fewer dense-core plaques than APP/PS1 mice with normal microglia. Our findings reveal that the TAM system is an essential mediator of microglial recognition and engulfment of amyloid plaques and that TAM-driven microglial phagocytosis does not inhibit, but rather promotes, dense-core plaque development.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
We declare that the main data supporting the findings of this study are available within the article and its Extended Data information files. Sequencing data were deposited in the Gene Expression Omnibus with the accession number GSE160523. Additional supporting raw data are available from the authors upon request. Source data are provided with this paper.
References
Lemke, G. Biology of the TAM receptors. Cold Spring Harb. Perspect. Biol. https://doi.org/10.1101/cshperspect.a009076 (2013).
Lu, Q. & Lemke, G. Homeostatic regulation of the immune system by receptor tyrosine kinases of the Tyro 3 family. Science 293, 306–311 (2001).
Lemke, G. How macrophages deal with death. Nat. Rev. Immunol. 19, 539–549 (2019).
Scott, R. S. et al. Phagocytosis and clearance of apoptotic cells is mediated by Mer. Nature 411, 207–211 (2001).
Zagórska, A., Través, P. G., Lew, E. D., Dransfield, I. & Lemke, G. Diversification of TAM receptor tyrosine kinase function. Nat. Immunol. 15, 920–928 (2014).
Rothlin, C. V., Ghosh, S., Zuniga, E. I., Oldstone, M. B. & Lemke, G. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell 131, 1124–1136 (2007).
Fourgeaud, L. et al. TAM receptors regulate multiple features of microglial physiology. Nature 532, 240–244 (2016).
Long, J. M. & Holtzman, D. M. Alzheimer disease: an update on pathobiology and treatment strategies. Cell 179, 312–339 (2019).
The ImmGen Consortium. Open-source ImmGen: mononuclear phagocytes. Nat. Immunol. 17, 741 (2016).
Lai, C. & Lemke, G. An extended family of protein-tyrosine kinase genes differentially expressed in the vertebrate nervous system. Neuron 6, 691–704 (1991).
Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290 (2017).
Yin, Z. et al. Immune hyperreactivity of Aβ plaque-associated microglia in Alzheimer’s disease. Neurobiol. Aging 55, 115–122 (2017).
Lew, E. D. et al. Differential TAM receptor–ligand–phospholipid interactions delimit differential TAM bioactivities. Elife 3, e03385 (2014).
Mattsson, N. et al. CSF protein biomarkers predicting longitudinal reduction of CSF β-amyloid42 in cognitively healthy elders. Transl. Psychiatry 3, e293 (2013).
Sainaghi, P. P. et al. Growth arrest specific 6 concentration is increased in the cerebrospinal fluid of patients with Alzheimer’s disease. J. Alzheimers Dis. 55, 59–65 (2017).
Jankowsky, J. L. et al. Mutant presenilins specifically elevate the levels of the 42 residue β-amyloid peptide in vivo: evidence for augmentation of a 42-specific γ secretase. Hum. Mol. Genet. 13, 159–170 (2004).
Jankowsky, J. L. et al. Co-expression of multiple transgenes in mouse CNS: a comparison of strategies. Biomol. Eng. 17, 157–165 (2001).
Rockenstein, E., Mallory, M., Mante, M., Sisk, A. & Masliaha, E. Early formation of mature amyloid-β protein deposits in a mutant APP transgenic model depends on levels of Aβ1–42. J. Neurosci. Res. 66, 573–582 (2001).
Labzin, L. I., Heneka, M. T. & Latz, E. Innate immunity and neurodegeneration. Annu Rev. Med. 69, 437–449 (2018).
Roy, E. R. et al. Type I interferon response drives neuroinflammation and synapse loss in Alzheimer disease. J. Clin. Invest. 130, 1912–1930 (2020).
Krasemann, S. et al. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 47, 566–581 (2017).
Ulland, T. K. & Colonna, M. TREM2 — a key player in microglial biology and Alzheimer disease. Nat. Rev. Neurol. 14, 667–675 (2018).
Bennett, M. L. et al. New tools for studying microglia in the mouse and human CNS. Proc. Natl Acad. Sci. USA 113, E1738–E1746 (2016).
Kim, D. K. et al. Deep proteome profiling of the hippocampus in the 5XFAD mouse model reveals biological process alterations and a novel biomarker of Alzheimer’s disease. Exp. Mol. Med. 51, 1–17 (2019).
Kim, Y. E., Chen, J., Langen, R. & Chan, J. R. Monitoring apoptosis and neuronal degeneration by real-time detection of phosphatidylserine externalization using a polarity-sensitive indicator of viability and apoptosis. Nat. Protoc. 5, 1396–1405 (2010).
Friedman, B. A. et al. Diverse brain myeloid expression profiles reveal distinct microglial activation states and aspects of Alzheimer’s disease not evident in mouse models. Cell Rep. 22, 832–847 (2018).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Gonzalez, H. & Pacheco, R. T-cell-mediated regulation of neuroinflammation involved in neurodegenerative diseases. J. Neuroinflammation 11, 201 (2014).
Spangenberg, E. et al. Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer’s disease model. Nat. Commun. 10, 3758 (2019).
Lu, Q. et al. Tyro-3 family receptors are essential regulators of mammalian spermatogenesis. Nature 398, 723–728 (1999).
Jung, S. et al. Analysis of fractaÿlkine receptor CX3CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell. Biol. 20, 4106–4114 (2000).
Klunk, W. E. et al. Imaging Aβ plaques in living transgenic mice with multiphoton microscopy and methoxy-X04, a systemically administered Congo red derivative. J. Neuropathol. Exp. Neurol. 61, 797–805 (2002).
Nimmerjahn, A., Kirchhoff, F. & Helmchen, F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308, 1314–1318 (2005).
Nimmerjahn, A. Two-photon imaging of microglia in the mouse cortex in vivo. Cold Spring Harb. Protoc. https://doi.org/10.1101/pdb.prot069294 (2012).
Wang, Y. et al. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell 160, 1061–1071 (2015).
Wang, Y. et al. TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J. Exp. Med. 213, 667–675 (2016).
Condello, C., Yuan, P., Schain, A. & Grutzendler, J. Microglia constitute a barrier that prevents neurotoxic protofibrillar Aβ42 hotspots around plaques. Nat. Commun. 6, 6176 (2015).
Hassiotis, S. et al. Lysosomal LAMP1 immunoreactivity exists in both diffuse and neuritic amyloid plaques in the human hippocampus. Eur. J. Neurosci. 47, 1043–1053 (2018).
Hu, X. et al. Transgenic mice overexpressing reticulon 3 develop neuritic abnormalities. EMBO J. 26, 2755–2767 (2007).
Greenberg, S. M. et al. Cerebral amyloid angiopathy and Alzheimer disease—one peptide, two pathways. Nat. Rev. Neurol. 16, 30–42 (2020).
Blanco-Suarez, E., Liu, T. F., Kopelevich, A. & Allen, N. J. Astrocyte-secreted chordin-like 1 drives synapse maturation and limits plasticity by increasing synaptic GluA2 AMPA receptors. Neuron 100, 1116–1132.e13 (2018).
Jay, T. R. et al. Disease progression-dependent effects of TREM2 deficiency in a mouse model of Alzheimer’s disease. J. Neurosci. 37, 637–647 (2017).
Jay, T. R. et al. TREM2 deficiency eliminates TREM2+ inflammatory macrophages and ameliorates pathology in Alzheimer’s disease mouse models. J. Exp. Med. 212, 287–295 (2015).
Yuan, P. et al. TREM2 haplodeficiency in mice and humans impairs the microglia barrier function leading to decreased amyloid compaction and severe axonal dystrophy. Neuron 90, 724–739 (2016).
Baik, S. H., Kang, S., Son, S. M. & Mook-Jung, I. Microglia contributes to plaque growth by cell death due to uptake of amyloid β in the brain of Alzheimer’s disease mouse model. Glia 64, 2274–2290 (2016).
Sosna, J. et al. Early long-term administration of the CSF1R inhibitor PLX3397 ablates microglia and reduces accumulation of intraneuronal amyloid, neuritic plaque deposition and pre-fibrillar oligomers in 5XFAD mouse model of Alzheimer’s disease. Mol. Neurodegener. 13, 11 (2018).
Fu, H. et al. Complement component C3 and complement receptor type 3 contribute to the phagocytosis and clearance of fibrillar Aβ by microglia. Glia 60, 993–1003 (2012).
Hu, X. et al. Amyloid seeds formed by cellular uptake, concentration, and aggregation of the amyloid-β peptide. Proc. Natl Acad. Sci. USA 106, 20324–20329 (2009).
Cambier, C. J., Falkow, S. & Ramakrishnan, L. Host evasion and exploitation schemes of Mycobacterium tuberculosis. Cell 159, 1497–1509 (2014).
Panza, F., Lozupone, M., Logroscino, G. & Imbimbo, B. P. A critical appraisal of amyloid-β-targeting therapies for Alzheimer disease. Nat. Rev. Neurol. 15, 73–88 (2019).
Hutter-Paier, B. et al. The ACAT inhibitor CP-113,818 markedly reduces amyloid pathology in a mouse model of Alzheimer’s disease. Neuron 44, 227–238 (2004).
Imbimbo, B. P. et al. CHF5074, a novel γ-secretase modulator, attenuates brain β-amyloid pathology and learning deficit in a mouse model of Alzheimer’s disease. Br. J. Pharmacol. 156, 982–993 (2009).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Bais, A. S. & Kostka, D. scds: computational annotation of doublets in single-cell RNA sequencing data. Bioinformatics 36, 1150–1158 (2020).
Schmidt, S. D., Mazzella, M. J., Nixon, R. A. & Mathews, P. M. Aβ measurement by enzyme-linked immunosorbent assay. Methods Mol. Biol. 849, 507–527 (2012).
Tufail, Y. et al. Phosphatidylserine exposure controls viral innate immune responses by microglia. Neuron 93, 574–586 (2017).
Koenigsknecht-Talboo, J. et al. Rapid microglial response around amyloid pathology after systemic anti-Aβ antibody administration in PDAPP mice. J. Neurosci. 28, 14156–14164 (2008).
Bolmont, T. et al. Dynamics of the microglial/amyloid interaction indicate a role in plaque maintenance. J. Neurosci. 28, 4283–4292 (2008).
Bernier, L. P. et al. Nanoscale surveillance of the brain by microglia via cAMP-regulated filopodia. Cell Rep. 27, 2895–2908 (2019).
Kitamura, T. et al. Island cells control temporal association memory. Science 343, 896–901 (2014).
Acknowledgements
We thank J. Hash for technical assistance, M. Mercken (Janssen Pharmaceuticals) for Aβ42 antibodies, R. Rissman and the ADRC at UCSD for AD brain sections, M. Shokhirev for advice on bioinformatic analyses, S. Parylak and F. Gage for advice on fear-conditioning assays and members of the Lemke laboratory and the Nomis Center for discussions. The research was supported by grants from the US National Institutes of Health (RF1 AG060748 and R01 AI101400 to G.L.; DP2 NS083038, R01 NS108034 and U01 NS103522 to A.N.; P30 AG062429 to the UCSD ADRC; and P30 CA014195 and S10-OD023689 to the Salk Institute), the Cure Alzheimer’s Fund and the Coins for Alzheimer’s Research Trust (to G.L.) and the Leona M. and Harry B. Helmsley Charitable Trust (to the Salk Institute); by Goeddel’s Chancellor’s, Marguerite Vogt and H. A. and Mary K. Chapman Charitable Trust graduate fellowships (to Y.H.) and by Anderson, Nomis and Sweden–America Foundation postdoctoral fellowships (to K.E.H.).
Author information
Authors and Affiliations
Contributions
Y.H. was an essential contributor to every aspect of the paper. K.E.H. made extensive contributions to experimental design and implementation. P.G.B. obtained and prepared tissue sections for immunostaining. C.O’C. contributed to the design and performance of flow cytometry. N.H. carried out scRNA-seq. L.H. performed clustering and bioinformatics for scRNA-seq analyses. A.N. designed and implemented methods for two-photon imaging and data analyses. G.L. conceived the project and designed experiments. All authors contributed to the writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Immunology thanks Cristoforo Comi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. L. A. Dempsey was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Expression of Axl, Mer, TMEM119, Trem2, and GFAP in plaque-burdened AD brains.
a, Axl protein (green, lower panels) is undetectable in WT (left) and APP/PS1 (right) microglia (Iba1, red, upper panels) in the 4mo cortex, prior to the appearance of plaques in APP/PS1 mice. n = 3 per genotype. b, Axl protein (green) in 15 mo APP41 mice (n = 3) is up-regulated in cortical microglia (Iba1, red) contacting Aβ plaques (6E10, white), and is also often concentrated in plaque centers, consistent with strong prior activation of Axl and subsequent cleavage of its ectodomain. (As is seen for other RTKs, robust activation of Axl results in nearly complete cleavage of the ectodomain from the cell surface.) Hoechst 33258 stains nuclei. c, Expression of Mer protein (green) in 15 mo APP41 mice is seen in all cortical microglia (Iba1, red), but is further up-regulated in microglia that invest Aβ plaques (6E10, white). d, Quantification of Mer up-regulation in Iba1+ plaque-associated microglia (PAM) versus non-plaque-associated microglia (NPAM) in APP/PS1 mice at 9.5 months. e, Axl (green) and Trem2 (red) are up-regulated in the same Iba1+ (white) microglia cell in the 9.5 mo APP/PS1 cortex. f, Expression of the homeostatic microglial marker TMEM119 (green) is lost in cortical microglia that surround plaques (6E10, white) in 15 mo APP41 mice, except for an occasional 1-2 cells at the center of plaques. g, This same TMEM119 down-regulation is seen in 15 mo APP/PS1 mice. TMEM119- microglia surrounding 6E10 + plaques are strongly Iba1+. h, The up-regulated Mer expression (green) seen in 15 mo APP41 mice is not in GFAP+ reactive astrocytes (red). (Activated S100b+ astrocytes are also negative for Mer expression by IHC7). Circles mark the position of Aβ plaques. Representative images obtained from immunostaining of N ≥ 3 sections from n = 3 mice of each genotype. Scale bars: 10 μm (a-c, e, h), 100 μm (f, g). Mann-Whitney test (d). For all supplementary figure panels, data are represented as mean ±1 STD.
Extended Data Fig. 2 Gas6 and PtdSer decoration of Aβ plaques.
a, Gas6 protein (green) decorates 6E10+ plaques (white) on sections of postmortem cortex from human patients with advanced (BRAAK stage 6) AD (left panels), but is not evident in the plaque-free cortex of cognitively normal age-matched controls (right panels). Representative images obtained from immunostaining of >3 sections from 3 individuals of each condition. b, Visualization of externalized PtdSer in 15 mo APP/PS1 cortex following stereotaxic injection of pSIVA (green, left panel). pSIVA binds to externalized PtdSer in the needle track of the injection, where cells are damaged and undergoing apoptosis, and also to the PtdSer associated with all 6E10+ amyloid plaques adjacent to the injection site (white, right panel). (c) A similar pSIVA injection in 15 mo WT mice labels only the needle track of the injection, since there are no plaques in these mice. (d) Airyscan super-resolution image of the juxtaposition of plaque-associated microglia (Iba1, red), Gas6 (green), and Aβ plaque (6E10, white) in 12 mo APP/PS1 mice (n = 5). Scale bars: 100 μm (a), 50 μm (b), 200 μm (c), 10 μm (d). n = 3 and 2 for APP/PS1 and WT control, respectively (b, c) from two independent experiments.
Extended Data Fig. 3 Transcriptomics of APP/PS1 and APP/PS1Axl−/−Mertk−/− microglia as quantified by single cell RNA-seq.
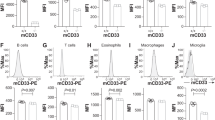
a, Sorting scheme for isolation of CD45+ single cells. FSC and SSC, forward and side scatter, respectively. A, area; W, width. b, Uniform manifold approximation and projection (UMAP) clustering of CD45+ cells sorted from 18 mo APP/PS1 (A/PS) and APP/PS1Axl−/−Mertk−/− (A/PS A/M−/−) cortices (combined) and annotated using the 18 marker genes in c. c, Violin plots of population log-transformed normalized expression of the indicated genes in the indicated cell types. Cell type key applies to b and c. d, Log-transformed normalized expression of Axl (left) and Mertk (right) mRNA in APP/PS1 cells within the microglial clusters defined in Fig. 3a. e, Heat map of the scaled expression levels of the indicated genes in individual cells within transcriptomic state (cluster) 0 and state 5 microglia in the APP/PS1 cortex at 18 mo. f, Comparative composite heat maps for the indicated genes across stages and transcriptomic states in 5xFAD and APP/PS1 mice, as quantified in this study (top five rows) and in Keren-Shaul et al.11 (bottom three rows), respectively. Values shown as z-scaled log-transformed normalized average of each group. g, Violin plots of the log-transformed normalized expression distribution of the indicated genes at transcriptomic state 5 in A/PS (black) versus A/PS A/M−/− (red) microglia, as determined by single cell RNA-seq (scRNA-seq). Dotted lines indicate mean. h, Mean expression level of the indicated cytokine and chemokine genes in state 5 versus state 0 microglia in A/PS (black) versus A/PS A/M−/− (red) microglia at 18 mo, as determined by scRNA-seq. i, Relative expression level of the indicated inflammatory regulator mRNAs in RNA isolated from total cortex of mice of the indicated genotypes at 12 mo, as determined by qRT-PCR. n = 3-6. Kruskal-Wallis test with Dunn’s multiple comparison test. Data are represented as mean ±1 STD.
Extended Data Fig. 4 APP/PS1Axl−/−Mertk−/− microglia are unresponsive to Aβ plaques.
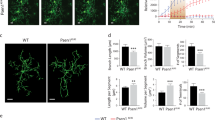
a, Distribution of distance of microglial cell body centroids, in 2 μm bins, from the edge of MX04-labeled Aβ plaques with diameters of 10-15 μm in APP/PS1 (gray) and APP/PS1Axl−/−Mertk−/− (red) cortex. Values obtained for 56 and 23 plaques from 3 and 4 mice for APP/PS1 and APP/PS1Axl−/−Mertk−/−, respectively. b, Number of imaged GFP+ primary processes per PAM (microglia < 5 μm from plaques) in APP/PS1 (gray, A/PS) and APP/PS1Axl−/−Mertk−/− (red, A/PS A/M−/−) cortex. c, Summed length of primary microglial processes per PAM in APP/PS1 (gray) and APP/PS1Axl−/−Mertk−/− (red) cortex. d, Process polarization ratio to nearest plaque per NPAM (microglia > 20 μm from plaques; see Materials and Methods) in APP/PS1 (gray) and APP/PS1Axl−/−Mertk−/− (red) cortex. e, Quantification of microglial cell density in the cortex of 16 mo APP/PS1 (gray) and APP/PS1Axl−/−Mertk−/− (red) mice for microglia 0-10 μm, 10-20 μm, and >20 μm from the edge of the nearest plaque. Data points are from 45-129 cells (PAM) investing 10-29 plaques (b, c), and 21-49 cells peripheral to 7-24 plaques (d) from n = 3 mice per genotype (b-d), Points in e represent 3-5 imaging volumes from 3 APP/PS and 4 APP/PS1Axl−/−Mertk−/− mice. Two-way ANOVA with Sidak’s multiple comparison test (a, e) and Mann-Whitney’s test (b-d), Data are represented as mean ±1 STD.
Extended Data Fig. 5 Expansive areas of plaque-associated dystrophic LAMP1+ membrane and poorly compacted plaques in the APP/PS1Axl−/−Mertk−/− brain.
A montage of 24 paired sections in both APP/PS1 (top six rows) and APP/PS1Axl−/−Mertk−/− (bottom six rows) cortex, each stained with antibodies to both LAMP1 (green) and 6E10 (white). Each 6E10 image is paired with (from the same section as) the LAMP image immediately below. This montage, which is a subset of the images used to generate the data in Fig. 5d, is composed of images taken from three different mice of each genotype at 12 months. Note that: (a) 6E10+ Aβ plaques are in general more compact and brightly stained in APP/PS1 mice and more diffuse and weakly stained in APP/PS1Axl−/−Mertk−/− mice; and (b) the area occupied by LAMP1+ membrane is in general much larger in APP/PS1Axl−/−Mertk−/− mice. Scale bars: 10 μm.
Extended Data Fig. 6 Accumulation of LAMP1+ dystrophic membrane and apoptotic cell debris in the APP/PS1Axl−/−Mertk−/− brain.
a, Quantification of LAMP1/6E10 area ratio as in Fig. 5d, but only for dense-core plaques (plaques with solid 6E10+ cores with areas > 100 μm2). b, Quantification of LAMP1/6E10 area ratio as in Fig. 5d, but only for diffuse plaques (plaques without solid 6E10+ cores with areas > 100 μm2). c, Quantification of RTN3/6E10 area ratio as in Fig. 5e, but only for dense-core plaques. d, Quantification of RTN3/6E10 area ratio as in Fig. 5e, but only for diffuse plaques. e, Quantification of the density of diffuse plaques (defined as above) expressed as a fraction of total plaques in the cortex of mice of the indicated genotypes at 12 mo. Data represent diffuse plaques quantified from N = 4-5 sections from n = 6 mice per group. (f) Representative example of cerebral amyloid angiopathy (CAA) in the cortex of a 15 mo APP/PS1Axl−/−Mertk−/− mouse. 6E10+ Aβ material (white) is evident within laminin+ blood vessels (green). Asterisk marks an Aβ plaque in the parenchyma. (g) Quantification (see Methods) of CAA in the somatosensory cortex of 15 mo APP/PS1Axl−/−Mertk−/− mice (A/PS A/M−/−) relative to APP/PS1 mice (A/PS). n = 4/group and measurements were averaged from N > 15 sections (spanning > 1.8mm3 of brain volume) per mouse. (h) cCasp3+ apoptotic debris (cyan, lower panels) accumulates around 6E10+ Aβ plaques (upper panels) in the APP/PS1Axl−/−Mertk−/− (right panels) but not the APP/PS1 (left panels) hippocampus at 12 mo. Images are representative of n = 3 mice per genotype from three independent experiments. Scale bars: 100 μm. Data are 18-47 (a), 67-78 (b), 30-43 (c) and 26-52 (d) plaques investigated from N ≥ 3 sections per mouse from n = 3 mice of each genotype from at least 3 independent replicates. Mann-Whitney test (a-e, g). Data are represented as mean ±1 STD.
Extended Data Fig. 7 TAM (Mer) signaling promotes dense-core Aβ plaque accumulation with functional consequences.
a, Thio S plaque density in APP/PS1 (gray) versus APP/PS1Axl−/−Mertk−/− (red) cortex for plaques of the indicated size at 12 months. b, Thio S+ plaque density (all plaque sizes) in APP/PS1 (gray) versus APP/PS1Axl−/−Mertk−/− (red) hippocampus at 12 months. c, Soluble Aβ42 levels quantified in APP/PS1 (gray) versus APP/PS1Axl−/−Mertk−/− (red) cortex and hippocampus at 4 and 12 mo, as indicated. n = 5-6 per genotype. d, Quantitative LI-COR western blot measurement of APP protein levels in the 12 mo cortex of 3 cohorts of mice (4 genotypes each cohort) of the indicated genotypes demonstrates no change in APP expression in APP/PS1 mice upon mutation of Axl and Mertk. Blots left and quantification right. e, ThioS+ plaque density (all plaque sizes) in APP/PS1 (gray) versus APP/PS1Mertk−/− (pink) cortex and hippocampus at 12 months. f, Thio S+ plaque density (all plaque sizes) in APP/PS1 (gray) versus APP/PS1Axl−/− (white) cortex and hippocampus at 12 months. Data points represent plaque density in n = 6-8 mice of the indicated genotypes averaged from N ≥ 5 cortical sections for each brain. Mann-Whitney test (a, c, d) and Student’s t-test (b, e, f). Data are represented as mean ±1 STD. g, TAM-mediated microglial recognition, phagocytosis, and consolidation of Aβ plaques. Microglial Axl and Mer are bridged to the PtdSer-rich dystrophic membranes of plaques via TAM ligands, whose amino-terminal and carboxy-terminal domains bind PtdSer and Axl/Mer, respectively3,26. Gas6 is shown, but a role for the Mer ligand Pros126 is not excluded. Engagement of the PtdSer-TAM ligand-TAM receptor complex activates the TAM tyrosine kinases (TK), which drives phagocytosis of forming plaque material. Internalized phagocytic cargo is eventually transferred to lysosomes, whose acidic interiors promote the aggregation of large, insoluble Aβ fibrils. Exocytosis or microglial death then delivers this aggregated material to growing dense-core plaques.
Extended Data Fig. 8 Functional consequences of TAM deletion in APP/PS1 mice.
a, Quantification of the 3D colocalization of the excitatory pre- and post-synaptic markers vGlut1 and PSD95 in the 15-mo hippocampus (CA1) (see Methods), as an index of synaptic connectivity. The previously documented decrease in co-localization of these markers in APP/PS1 mice is not altered by the combined mutation of Axl and Mertk. Stack size is 85 x 85 x3 μm3 per image, averaged 3 images per CA1 section across 3-5 sections per mouse. Data points represent synaptic density index in n = 3 mice of the indicated genotypes. b, Acquisition of association between a 30 s auditory tone and a subsequent co-terminal 2 s 0.5 mA foot shock, expressed as percent time immobile (% freezing) during the indicated intervals, over three successive trial intervals in 15 mo mice of the indicated genotypes (see Methods). c, Data in b plotted for the indicated genotypes with the indicated statistical significance per interval. d, Contextual fear memory as assayed by percent of a 3 minute interval in which mice of the indicated genotypes were immobile (% freezing) when returned to the same testing cage 24 h after the fear acquisition trials of b (see Methods). A cohort of group-housed male mice (n = 12-20/group) were used in the behavioral assay. Data points in b represent the mean % freezing of each group in the interval duration immediately prior to the point. Each data point in d is the % freezing of one animal in the duration of the testing period. Kruskal Wallis test followed by Dunn’s multiple comparison test (a) and Mann-Whitney test (c, d). Data are represented as mean ±1 STD.
Supplementary information
Supplementary Video 1
Microglia show reduced interaction with amyloid plaques in somatosensory cortical layer 1 of 16-mo APP/PS1Axl−/−Mertk−/− as compared to APP/PS1 mice. Left, example two-photon time-lapse recording showing microglia (GFP signal, white) and amyloid plaques (MX04 signal, red) in somatosensory cortical layer 1 of an anesthetized APP/PS1Cx3cr1GFP/+ mouse. Each image is a maximum-intensity projection of a 70-μm-thick z-stack (1.2 μm axial step size) recorded every ~93 s at depths between 100 μm and 30 μm below the pia. Right, example two-photon time-lapse recording showing microglia (GFP signal, white) and amyloid plaques (MX04 signal, red) in somatosensory cortical layer 1 of an age- and sex-matched anesthetized APP/PS1Axl−/−Mertk−/−Cx3cr1GFP/+ mouse. Each image is a maximum-intensity projection of a 70-μm-thick z-stack (1.2 μm axial step size) recorded every ~93 s at depths between 94 μm and 24 μm below the pia. Elapsed time is indicated in the upper right corner. Amyloid plaques are closely encapsulated by highly polarized microglia in APP/PS1 but not APP/PS1Axl−/−Mertk−/− mice. Additionally, microglia near plaques typically show fewer and less-elaborate processes, higher process polarization and reduced process motility in APP/PS1 as compared to APP/PS1Axl−/−Mertk−/− mice. Supplementary Videos 1 and 2 were prepared using FIJI and custom-written scripts in Igor Pro (v.6.04; WaveMetrics). Scale bar, 30 μm.
Supplementary Video 2
Microglia show reduced interaction with amyloid plaques in somatosensory cortical layers 1/2 of 16-mo APP/PS1Axl−/−Mertk−/− as compared to APP/PS1 mice. Left, example two-photon time-lapse recording showing microglia (GFP signal, white) and amyloid plaques (MX04 signal, red) in somatosensory cortical layers 1/2 of an anesthetized APP/PS1Cx3cr1GFP/+ mouse. Each image is a maximum-intensity projection of a 70-μm-thick z-stack (1.2 μm axial step size) recorded every ~93 s at depths between 148 μm and 78 μm below the pia. Right, example two-photon time-lapse recording showing microglia (GFP signal, white) and amyloid plaques (MX04 signal, red) in somatosensory cortical layers 1/2 of an age-and sex-matched anesthetized APP/PS1Axl−/−Mertk−/−Cx3cr1GFP/+ mouse. Each image is a maximum-intensity projection of a 70-μm-thick z-stack (1.2 μm axial step size) recorded every ~93 s at depths between 135 μm and 65 μm below the pia. Elapsed time is indicated in the upper right corner. Amyloid plaques are closely encapsulated by highly polarized microglia in APP/PS1 but not APP/PS1Axl−/−Mertk−/− mice. Additionally, microglia near plaques typically show fewer and less-elaborate processes, higher process polarization and reduced process motility in APP/PS1 as compared to APP/PS1Axl−/−Mertk−/− mice. Scale bar, 30μm.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 7
Unprocessed immunoblots.
Source Data Extended Data Fig. 8
Statistical source data.
Rights and permissions
About this article
Cite this article
Huang, Y., Happonen, K.E., Burrola, P.G. et al. Microglia use TAM receptors to detect and engulf amyloid β plaques. Nat Immunol 22, 586–594 (2021). https://doi.org/10.1038/s41590-021-00913-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-021-00913-5
This article is cited by
-
Modulation of hippocampal protein expression by a brain penetrant biologic TNF-α inhibitor in the 3xTg Alzheimer’s disease mice
Journal of Translational Medicine (2024)
-
Altered amyloid-β structure markedly reduces gliosis in the brain of mice harboring the Uppsala APP deletion
Acta Neuropathologica Communications (2024)
-
Taming microglia: the promise of engineered microglia in treating neurological diseases
Journal of Neuroinflammation (2024)
-
Exercise mimetics: a novel strategy to combat neuroinflammation and Alzheimer’s disease
Journal of Neuroinflammation (2024)
-
Microbiota–gut–brain axis and its therapeutic applications in neurodegenerative diseases
Signal Transduction and Targeted Therapy (2024)