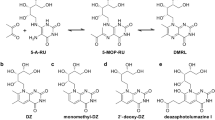
Abstract
Mucosal-associated invariant T (MAIT) cells are activated by microbial riboflavin-based metabolite antigens when presented by MR1. How modifications to the potent antigen 5-OP-RU affect presentation by MR1 and MAIT cell activation remains unclear. Here we design 20 derivatives, termed altered metabolite ligands (AMLs), to dissect the impact of different antigen components on the human MAIT–MR1 axis. Analysis of 11 crystal structures of MAIT T cell antigen receptor (TCR)–MR1–AML ternary complexes, along with biochemical and functional assays, shows that MR1 cell-surface upregulation is influenced by ribityl and non-ribityl components of the ligand and the hydrophobicity of the MR1–AML interface. The polar ribityl chain of the AML strongly influences MAIT cell activation potency through dynamic compensatory interactions within a MAIT TCR–MR1–AML interaction triad. We define the basis by which the MAIT TCR can differentially recognize AMLs, thereby providing insight into MAIT cell antigen specificity and potency.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon request. The atomic coordinates and structure factors of MAIT A-F7 TCR–MR1–AML ternary complexess with AMLs 5-OP-RU, 5′-OH-pentyl-5-OP-U, 4′-D-5-OP-RU, 5′-D-5-OP-RU, 2′-OH-ethyl-5-OP-U, ribityl-less analog, 4′-OH-butyl-5-OP-U, 3′-OH-propyl-5-OP-U, JYM72, 3′-D-5-OP-RU and 2′-D-5-OP-RU have been deposited in the Protein Data Bank under accession codes 6PUC, 6PUD, 6PUE, 6PUF, 6PUG, 6PUH, 6PUI, 6PUJ, 6PUK, 6PUL and 6PUM, respectively.
References
Rossjohn, J. et al. T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 33, 169–200 (2015).
La Gruta, N. L., Gras, S., Daley, S. R., Thomas, P. G. & Rossjohn, J. Understanding the drivers of MHC restriction of T cell receptors. Nat. Rev. Immunol. 18, 467–478 (2018).
Sloan-Lancaster, J. & Allen, P. M. Altered peptide ligand-induced partial T cell activation: molecular mechanisms and role in T cell biology. Annu. Rev. Immunol. 14, 1–27 (1996).
Evavold, B. D., Sloan-Lancaster, J. & Allen, P. M. Tickling the TCR: selective T-cell functions stimulated by altered peptide ligands. Immunol. Today 14, 602–609 (1993).
Van Rhijn, I., Godfrey, D. I., Rossjohn, J. & Moody, D. B. Lipid and small-molecule display by CD1 and MR1. Nat. Rev. Immunol. 15, 643–654 (2015).
Borg, N. A. et al. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature 448, 44–49 (2007).
Wun, K. S. et al. A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells. Immunity 34, 327–339 (2011).
De Libero, G., Lau, S.-Y. & Mori, L. Phosphoantigen presentation to TCR γδ cells, a conundrum getting less gray zones. Front. Immunol. 5, 679 (2015).
Kjer-Nielsen, L. et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491, 717–723 (2012).
Corbett, A. J. et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 509, 361–365 (2014).
Treiner, E. et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 422, 164–169 (2003).
Le Bourhis, L. et al. Mucosal-associated invariant T cells: unconventional development and function. Trends Immunol. 32, 212–218 (2011).
Reantragoon, R. et al. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J. Exp. Med. 210, 2305–2320 (2013).
Toubal, A., Nel, I., Lotersztajn, S. & Lehuen, A. Mucosal-associated invariant T cells and disease. Nat. Rev. Immunol. 19, 643–657 (2019).
Eckle, S. B. et al. A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. J. Exp. Med. 211, 1585–1600 (2014).
Gherardin, N. A. et al. Diversity of T cells restricted by the MHC class I-related molecule MR1 facilitates differential antigen recognition. Immunity 44, 32–45 (2016).
Lepore, M. et al. Parallel T-cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRβ repertoire. Nat. Commun. 5, 3866 (2014).
Patel, O. et al. Recognition of vitamin B metabolites by mucosal-associated invariant T cells. Nat. Commun. 4, 2142 (2013).
Keller, A. N. et al. Drugs and drug-like molecules can modulate the function of mucosal-associated invariant T cells. Nat. Immunol. 18, 402–411 (2017).
Le Nours, J. et al. A class of γδ T cell receptors recognize the underside of the antigen-presenting molecule MR1. Science 366, 1522–1527 (2019).
Keller, A. N., Corbett, A. J., Wubben, J. M., McCluskey, J. & Rossjohn, J. MAIT cells and MR1–antigen recognition. Curr. Opin. Immunol. 46, 66–74 (2017).
Awad, W., Le Nours, J., Kjer-Nielsen, L., McCluskey, J. & Rossjohn, J. Mucosal-associated invariant T cell receptor recognition of small molecules presented by MR1. Immunol. Cell Biol. 96, 588–597 (2018).
Ler, G. J. M. et al. Computer modelling and synthesis of deoxy and monohydroxy analogues of a ribitylaminouracil bacterial metabolite that potently activates human T cells. Chem. Eur. J. 25, 15594–15608 (2019).
Mak, J. Y. W. et al. Stabilizing short-lived Schiff base derivatives of 5-aminouracils that activate mucosal-associated invariant T cells. Nat. Commun. 8, 14599 (2017).
Reantragoon, R. et al. Structural insight into MR1-mediated recognition of the mucosal associated invariant T cell receptor. J. Exp. Med. 209, 761–774 (2012).
McWilliam, H. E. G. et al. The intracellular pathway for the presentation of vitamin B-related antigens by the antigen-presenting molecule MR1. Nat. Immunol. 17, 531–537 (2016).
Burrows, S. R. et al. Hard wiring of T cell receptor specificity for the major histocompatibility complex is underpinned by TCR adaptability. Proc. Natl Acad. Sci. USA 107, 10608–10613 (2010).
Tilloy, F. et al. An invariant T cell receptor α chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted α/β T cell subpopulation in mammals. J. Exp. Med. 189, 1907–1921 (1999).
Eckle, S. B. et al. Recognition of vitamin B precursors and byproducts by mucosal associated invariant T cells. J. Biol. Chem. 290, 30204–30211 (2015).
Harriff, M. J. et al. MR1 displays the microbial metabolome driving selective MR1-restricted T cell receptor usage. Science Immunol. 3, eaao2556 (2018).
Wang, H. et al. MAIT cells protect against pulmonary Legionella longbeachae infection. Nat. Commun. 9, 3350 (2018).
Wang, H. et al. IL-23 costimulates antigen-specific MAIT cell activation and enables vaccination against bacterial infection. Science Immunol. 4, eaaw0402 (2019).
Godfrey, D. I., Le Nours, J., Andrews, D. M., Uldrich, A. P. & Rossjohn, J. Unconventional T cell targets for cancer immunotherapy. Immunity 48, 453–473 (2018).
Crowther, M. D. et al. Genome-wide CRISPR–Cas9 screening reveals ubiquitous T cell cancer targeting via the monomorphic MHC class I-related protein MR1. Nat. Immunol. 21, 178–185 (2020).
Hwang, D. R., Helquist, P. & Shekhani, M. S. Total synthesis of (+)-sparsomycin. Approaches using cysteine and serine inversion. J. Org. Chem. 50, 1264–1271 (1985).
Huang, S. et al. Evidence for MR1 antigen presentation to mucosal-associated invariant T cells. J. Biol. Chem. 280, 21183–21193 (2005).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
McCoy, A. J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Acknowledgements
We thank the staff at the National Synchrotron for assistance with data collection and the staff at the Monash Macromolecular Crystallization Facility and the Doherty Flow Cytometry Facility. This research was undertaken in part by using the MX2 beamline at the Australian Synchrotron, part of ANSTO, and made use of the Australian Cancer Research Foundation (ACRF) detector. This work was supported by the Australian National Health and Medical Research Council (NHMRC; 1125493 and 1113293), the Australian Research Council (ARC) Centre of Excellence in Advanced Molecular Imaging (CE140100011) and The University of Queensland (UQECR1834385). D.P.F. was an NHMRC Senior Principal Research Fellow (1027369 and 1117017), S.E. is an ARC DECRA Fellow (DE170100407), A.J.C. (FT160100083) and J.L.N. (FT160100074) are ARC Future Fellows and J.R. is an Australian ARC Laureate Fellow (FL160100049).
Author information
Authors and Affiliations
Contributions
W.A. and G.J.M.L. are joint first authors and designed, performed and analyzed data from the experiments; W.A. co-wrote the paper. W.X., A.N.K., J.Y.W.M., X.Y.L., L.L., S.B.G.E., J.L.N. and J.M. performed experiments, provided key reagents, analyzed data and/or provided intellectual input; A.J.C., D.P.F. and J.R. are joint senior and corresponding authors.
Corresponding authors
Ethics declarations
Competing interests
A.J.C., S.B.G.E., J.Y.W.M., L.L., D.P.F., J.R. and J.M. are inventors on patents describing MR1 tetramers and MR1–ligand complexes.
Additional information
Peer review information Zoltan Fehervari was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Thermostability of soluble MR1-AML by fluorescence-based thermal shift assay.
a, Shown is baseline-corrected, normalized emission at 610 nm plotted against temperature using Boltzmann curve-fits. Data show mean ± S.E.M from three independent experiments (n = 3). The half maximum melt point (Tm50) is indicated as a dashed line. Displayed is a representative of three independent experiments yielding similar results. b, Table summarized the Tm50 of MR1-AMLs.
Extended Data Fig. 2 Activation of Jurkat.MAIT reporter cell lines and PBMC MAIT cells.
(a, b): Activation of Jurkat.MAIT reporter cells expressing TRBV6-1, TRBV6-4 and TRBV20 TCRs, as well as Jurkat.LC13 (HLA-B8-EBV peptide specific non-MAIT control) by 5-OP-RU, mono-deoxy analogues and stabilised AML (a) or mono-hydroxy analogues (b), at 10 μM for 16 hr in co-culture with C1R.MR1 cells as antigen presenting cells. FLR; Epstein-Barr viral peptide FLRGRAYGL, 285 nM. Data show mean ± S.E.M from n = 3 independent experiments. The AMLs variably activated the Jurkat.MAIT reporter cell lines, but not Jurkat cells expressing a non-MAIT TCR (Jurkat.LC13), revealing no TCR-independent activation by the AMLs. (c, d): Activation of PBMC MAIT cells (defined as CD3+CD161+MR1-5-OP-RU tetramer+). Representative plots (c) and Percentage of MAIT cells within human PBMCs expressing TNF and IFN-γ (d) after overnight incubation with 5-OP-RU or AML at indicated concentrations. Data show mean ± S.E.M. from n = 3 independent experiments on healthy blood donors. Gating strategy is shown in Extended Data Fig. 4.
Extended Data Fig. 3 Surface plasmon resonance (SPR) steady-state affinity measurements.
The binding affinity of MR1-AMLs versus two TCRs (a) TRAV1-2-TRBV6-1 and (b) TRAV1-2-TRBV6-4. SPR runs were conducted as duplicate (n = 2) using various proteins batches. The SPR sensograms, equilibrium curves and steady state KD values (µM) were prepared in GraphPad Prism 7.
Extended Data Fig. 4 Gating panel and plots for tetramer titration and PBMC MAIT cell activation assay.
a, gating strategy for PBMC MR1-AML-tetramer staining shown in Fig. 4a and for PBMC activation assay (Extended Data Fig. 2c, d). Cells were gated sequentially (from left to right) for lymphocytes (FSC-H vs SSC-H), single cells (FSC-H vs FSC-A), live T cells (CD3+7AAD−), and MAIT cells (CD161+MR1-AML-tetramer+). b, gating strategy for PBMC MR1-AML-tetramer co-staining (Fig. 4b, c). Cells were gated sequentially for lymphocytes (FSC-H vs SSC-H), single cells (FSC-H vs FSC-A), live T cells (CD3+7AAD−), and CD161hi cells. c, Percentage of MR1-AML+ MAIT cells that do not stain with MR1-5-OP-RU-tetramer. Individual values and mean ± SEM of 3 healthy donors (relates to Fig. 4b, c).
Extended Data Fig. 5 Final electron density maps of AML ligands.
The 2Fo-Fc map (at 1 sigma) of the AMLs that highlight the unambiguous positions of the AMLs in MR1 pocket. Working density map of 5-OP-RU a, JYM72 b, Ribityl-less analogue c, 5′-D−5-OP-RU d, 5′-OH-pentyl-5-OP-U e, 4′-D-5-OP-RU f, 4′-OH-butyl-5-OP-U g, 3′-D-5-OP-RU h, 3′-OH-propyl-5-OP-U i, 2′-D-5-OP-RU j, and 2′-OH-ethyl-5-OP-U AMLs k. Colours of the ligands are consistent with Fig. 5 and the density maps were shown as blue mesh.
Extended Data Fig. 6 AML interactions within the MR1 cleft.
a, superimposition of all TCR-MR1-AMLs. b, the top view of the Ag-binding cleft of the respective MR1 molecule displaying similar docking of the TCR CDR loops. (c–m) structures showing MR1 residues (white) that directly contacted: 5-OP-RU c, JYM72 d, Ribityl-less e, 5′-D-5-OP-RU f, 5′-OH-pentyl-5-OP-U g, 4′-D-5-OP-RU h, 4′-OH-butyl-5-OP-U i, 3′-D-5-OP-RU j, 3′-OH-propyl-5-OP-U k, 2′-D-5-OP-RU l, and 2′-OH-ethyl-5-OP-U AMLs m. Colours of the ligands are consistent with Fig. 5 and water molecules were shown as red spheres.
Extended Data Fig. 7 Conservation of “Interaction triad” among the typical MAIT TRAV1-2+ TCR-MR1-5-OP-RU ternary structures.
The variations in CDR3β loops manifested in differing loop recognition to 5-OP-RU via direct or water based interactions, yet still exhibited similar TCR binding affinities (~ 1 to 9 μM) and activation potency. (a-e) the binding affinities and the interactions between MR1 presenting 5-OP-RU and the CDR3α loop (light blue) and CDR3β loop (pink) of various TRAV1-2/TRBV6-1 TCRs carrying variable CDR3β loop including A-F7 a, B-B10 (PDB; 4PJA) b, C-C10 (PDB; 4PJC) c, B-F3-C1(PDB; 4PJB) d, and C-A11 (PDB; 4PJD) TCRs e. (f-i) the impact of usage of various TRAJ and TRBV genes on the complex interfaces between MR1-5-OP-RU molecule and the M33.64 (TRAJ33/TRBV6-4, PDB; 5D5M) f, #6 (TRAJ33/TRBV6-4, PDB; 4PJ7) g, C-F7 (TRAJ33/TRBV20, PDB; 4PJ8) h, and #4 (TRAJ20/TRBV6-4, PDB; 4PJ9) TCRs i.
Extended Data Fig. 8 The flexibility in the TCR-MR1-AML complexes.
(a–d) AML and MR1 flexibility in the TCR-MR1-AML complex in a simulated water environment. a, Measurement of ligand root mean squared fluctuations (RMSF) throughout 100 ns molecular dynamics simulations in water, starting from ternary crystal structures of the deoxy-AMLs. b, RMSF of the sidechains of selected MR1 α2 helix surface residues (single amino acid code) calculated throughout 100 ns molecular dynamics simulations in water, starting from the ternary crystal structures of the deoxy-AMLs. c, Pearson correlation analysis between the averaged RMSF of ligand heavy atoms, experimentally measured ligand binding affinity from SPR measurements of MAIT A-F7 TCR-MR1-AML ternary complexes (blue), and relative MAIT A-F7 TCR activation at a ligand concentration of 10 μM (red). d, Pearson correlation analysis between the averaged sidechain RMSF of MR1 α2 helix surface residues, the mean of the SPR binding affinity of MAIT A-F7 TCR-MR1-AML ternary complex (blue), and relative MAIT A-F7 TCR activation at a ligand concentration of 10 μM (red).
Supplementary information
Supplementary Tables
Supplementary Tables 1–3
Supplementary Video 1
Molecular movie of 100-ns simulation in water of 5-OP-RU-bound MR1–TCR ternary crystal structure. MR1 residue Tyr62 and Tyr95TCR3α interchangeably interact with and compete for the 2′-OH group. Tyr95TCR3α and MR1 residue Tyr152 contact one another and compete with both Tyr95TCR3α…2′-OH and MR1 residue Tyr152…5′-OH, while the 3′-OH and 4′-OH largely interact with Arg9 of MR1. The 3′-OH group remains hydrogen bonded to uracil N1H through most of the simulation. Carbon atoms are colored gray for ligand, orange for MR1 residues and cyan for Y95TCR3α. Hydrogen bonds are shown as black dashed sticks.
Supplementary Video 2
Molecular movie of 100-ns simulation in water of 2′-D-5-OP-RU-bound MR1–TCR ternary crystal structure. For the 2′-D-5-OP-RU analog, the 5′-hydroxyl group spends most of its time contacting Tyr152 and only a short time hydrogen bonding with Tyr95TCR3α. RMSF data suggest that the 2′-D-5-OP-RU AML is the most flexible compound at its 4′- and 5′-OH groups, while its 3′-OH group maintains the hydrogen bond with N1H of uracil. Carbon atoms are colored dark blue for ligand, orange for MR1 residues and cyan for Y95TCR3α. Hydrogen bonds are shown as black dashed sticks.
Supplementary Video 3
Molecular movie of 100-ns simulation in water of 3′-D-5-OP-RU-bound MR1–TCR ternary complex crystal structure. 3′-D-5-OP-RU AML formed a different intramolecular hydrogen bond between the 4′-OH group and uracil N1H that was maintained through the simulation. This substantially reduced flexibility at the adjacent 5′-OH and 2′-OH groups. The 4′- and 5′-OH groups faced toward MR1 and did not contact TCR3β, while Tyr62 of MR1 now competed effectively with Tyr95TCR3α for the 2′-OH group. Tyr95TCR3α mostly contacted Tyr152 of MR1 during the simulation. Carbon atoms are colored black for ligand, orange for MR1 residues and cyan for Y95TCR3α. Hydrogen bonds are shown as black dashed sticks.
Supplementary Video 4
Molecular movie of 100-ns simulation in water of 4′-D-5-OP-RU-bound MR1–TCR ternary complex crystal structure. The 4′-D-5-OP-RU AML was similar to 5-OP-RU, with the 2′-OH of the 2′-D-5-OP-RU ligand dividing its time between hydrogen bonding to MR1 residue Tyr62 and Tyr95TCR3α, with competition for Tyr95TCR3α from MR1 residue Tyr152. Carbon atoms are colored green for ligand, orange for MR1 residues, cyan for Y95TCR3α and purple for E99TCR3β. Hydrogen bonds are shown as black dashed sticks.
Supplementary Video 5
Molecular movie of 100-ns simulation in water of 5′-D-5-OP-RU-bound MR1–TCR ternary complex crystal structure. The 5′-D-5-OP-RU AML was similar to 5-OP-RU. There was little flexibility in the 5′-D-5-OP-RU ligand through the simulation. Carbon atoms are colored magenta for ligand, orange for MR1 residues and cyan for Y95TCR3α. Hydrogen bonds are shown as black dashed sticks.
Rights and permissions
About this article
Cite this article
Awad, W., Ler, G.J.M., Xu, W. et al. The molecular basis underpinning the potency and specificity of MAIT cell antigens. Nat Immunol 21, 400–411 (2020). https://doi.org/10.1038/s41590-020-0616-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-020-0616-6
This article is cited by
-
MR1 antigen presentation to MAIT cells and other MR1-restricted T cells
Nature Reviews Immunology (2024)
-
Unconventional immune cells in the gut mucosal barrier: regulation by symbiotic microbiota
Experimental & Molecular Medicine (2023)
-
Mucosal-associated invariant T cells display both pathogenic and protective roles in patients with inflammatory bowel diseases
Amino Acids (2023)
-
Deaza-modification of MR1 ligands modulates recognition by MR1-restricted T cells
Scientific Reports (2022)
-
Single-cell sequencing resolves the landscape of immune cells and regulatory mechanisms in HIV-infected immune non-responders
Cell Death & Disease (2022)