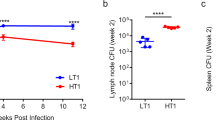
Abstract
Although mouse infection models have been extensively used to study the host response to Mycobacterium tuberculosis, their validity in revealing determinants of human tuberculosis (TB) resistance and disease progression has been heavily debated. Here, we show that the modular transcriptional signature in the blood of susceptible mice infected with a clinical isolate of M. tuberculosis resembles that of active human TB disease, with dominance of a type I interferon response and neutrophil activation and recruitment, together with a loss in B lymphocyte, natural killer and T cell effector responses. In addition, resistant but not susceptible strains of mice show increased lung B cell, natural killer and T cell effector responses in the lung upon infection. Notably, the blood signature of active disease shared by mice and humans is also evident in latent TB progressors before diagnosis, suggesting that these responses both predict and contribute to the pathogenesis of progressive M. tuberculosis infection.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The materials, data and any associated protocols that support the findings of this study are available from the corresponding author upon request. The RNA-seq datasets have been deposited in the NCBI Gene Expression Omnibus database with the primary accession number GSE140945 (TB mouse blood and lung). Publicly available datasets used in this study include GSE107995 (human TB datasets from Singhania et al.16) and GSE79362 (human TB dataset from Zak et al.38).
References
WHO. Global Tuberculosis Report 2017 (World Health Organization, 2017).
Dowdy, D. W., Basu, S. & Andrews, J. R. Is passive diagnosis enough? The impact of subclinical disease on diagnostic strategies for tuberculosis. Am. J. Resp. Crit. Care Med. 187, 543–551 (2013).
Casanova, J. L. & Abel, L. Genetic dissection of immunity to mycobacteria: the human model. Annu. Rev. Immunol. 20, 581–620 (2002).
Cooper, A. M., Mayer-Barber, K. D. & Sher, A. Role of innate cytokines in mycobacterial infection. Mucosal Immunol. 4, 252–260 (2011).
Flynn, J. L. & Chan, J. Immunology of tuberculosis. Annu. Rev. Immunol. 19, 93–129 (2001).
O’Garra, A. et al. The immune response in tuberculosis. Annu. Rev. Immunol. 31, 475–527 (2013).
Mayer-Barber, K. D. et al. Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk. Nature 511, 99–103 (2014).
Berry, M. P. et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 466, 973–977 (2010).
Blankley, S. et al. A 380-gene meta-signature of active tuberculosis compared with healthy controls. Eur. Resp. J. 47, 1873–1876 (2016).
Blankley, S. et al. The transcriptional signature of active tuberculosis reflects symptom status in extra-pulmonary and pulmonary tuberculosis. PloS One 11, e0162220 (2016).
Bloom, C. I. et al. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PloS One 8, e70630 (2013).
Joosten, S. A., Fletcher, H. A. & Ottenhoff, T. H. A helicopter perspective on TB biomarkers: pathway and process based analysis of gene expression data provides new insight into TB pathogenesis. PloS One 8, e73230 (2013).
Kaforou, M. et al. Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case-control study. PLoS Med. 10, e1001538 (2013).
Maertzdorf, J. et al. Human gene expression profiles of susceptibility and resistance in tuberculosis. Genes Immun. 12, 15–22 (2011).
Scriba, T. J. et al. Sequential inflammatory processes define human progression from M. tuberculosis infection to tuberculosis disease. PLoS Pathog. 13, e1006687 (2017).
Singhania, A. et al. A modular transcriptional signature identifies phenotypic heterogeneity of human tuberculosis infection. Nat. Commun. 9, 2308 (2018).
Behar, S. M., Divangahi, M. & Remold, H. G. Evasion of innate immunity by Mycobacterium tuberculosis: is death an exit strategy? Nat. Rev. Microbiol. 8, 668–674 (2010).
Moreira-Teixeira, L., Mayer-Barber, K., Sher, A. & O’Garra, A. Type I interferons in tuberculosis: foe and occasionally friend. J. Exp. Med. 215, 1273–1285 (2018).
Carmona, J. et al. Mycobacterium tuberculosis strains are differentially recognized by TLRs with an impact on the immune response. PloS One 8, e67277 (2013).
Dorhoi, A. et al. Type I IFN signaling triggers immunopathology in tuberculosis-susceptible mice by modulating lung phagocyte dynamics. Eur. J. Immunol. 44, 2380–2393 (2014).
Manca, C. et al. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-α/β. Proc. Natl Acad. Sci. USA 98, 5752–5757 (2001).
Manca, C. et al. Hypervirulent M. tuberculosis W/Beijing strains upregulate type I IFNs and increase expression of negative regulators of the Jak–Stat pathway. J. Interferon Cytokine Res. 25, 694–701 (2005).
Ordway, D. et al. The hypervirulent Mycobacterium tuberculosis strain HN878 induces a potent TH1 response followed by rapid down-regulation. J. Immunol. 179, 522–531 (2007).
McNab, F. W. et al. TPL-2-ERK1/2 signaling promotes host resistance against intracellular bacterial infection by negative regulation of type I IFN production. J. Immunol. 191, 1732–1743 (2013).
Bogunovic, D. et al. Mycobacterial disease and impaired IFN-γ immunity in humans with inherited ISG15 deficiency. Science 337, 1684–1688 (2012).
Antonelli, L. R. et al. Intranasal poly-IC treatment exacerbates tuberculosis in mice through the pulmonary recruitment of a pathogen-permissive monocyte/macrophage population. J. Clin. Invest. 120, 1674–1682 (2010).
Redford, P. S. et al. Influenza A virus impairs control of Mycobacterium tuberculosis coinfection through a type I interferon receptor-dependent pathway. J. Infect. Dis. 209, 270–274 (2014).
Barry, C. E. III et al. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat. Rev. Microbiol. 7, 845–855 (2009).
Kramnik, I. & Beamer, G. Mouse models of human TB pathology: roles in the analysis of necrosis and the development of host-directed therapies. Semin. Immunopathol. 38, 221–237 (2016).
Domaszewska, T. et al. Concordant and discordant gene expression patterns in mouse strains identify best-fit animal model for human tuberculosis. Sci. Rep. 7, 12094 (2017).
Singhania, A. et al. Transcriptional profiling unveils type I and II interferon networks in blood and tissues across diseases. Nat. Commun. 10, 2887 (2019).
Ley, K. et al. Neutrophils: new insights and open questions. Sci. Immunol. 3, eaat4579 (2018).
Dorhoi, A. et al. The adaptor molecule CARD9 is essential for tuberculosis control. J. Exp. Med. 207, 777–792 (2010).
Eruslanov, E. B. et al. Neutrophil responses to Mycobacterium tuberculosis infection in genetically susceptible and resistant mice. Infect. Immun. 73, 1744–1753 (2005).
Nandi, B. & Behar, S. M. Regulation of neutrophils by interferon-γ limits lung inflammation during tuberculosis infection. J. Exp. Med. 208, 2251–2262 (2011).
Achkar, J. M., Chan, J. & Casadevall, A. B cells and antibodies in the defense against Mycobacterium tuberculosis infection. Immunol. Rev. 264, 167–181 (2015).
Lu, L. L. et al. A functional role for antibodies in tuberculosis. Cell 167, 433–443 (2016).
Zak, D. E. et al. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet 387, 2312–2322 (2016).
Feng, C. G. et al. NK cell-derived IFN-γ differentially regulates innate resistance and neutrophil response in T cell-deficient hosts infected with Mycobacterium tuberculosis. J. Immunol. 177, 7086–7093 (2006).
Chowdhury, R. R. et al. A multi-cohort study of the immune factors associated with M. tuberculosis infection outcomes. Nature 560, 644–648 (2018).
Aly, S. et al. Oxygen status of lung granulomas in Mycobacterium tuberculosis-infected mice. J. Pathol. 210, 298–305 (2006).
Gopal, R. et al. S100A8/A9 proteins mediate neutrophilic inflammation and lung pathology during tuberculosis. Am. J. Resp. Crit. Care Med. 188, 1137–1146 (2013).
Eum, S.-Y. et al. Neutrophils are the predominant infected phagocytic cells in the airways of patients with active pulmonary TB. Chest 137, 122–128 (2010).
Verma, R. et al. A novel high sensitivity bacteriophage-based assay identifies low level M. tuberculosis bacteraemia in immunocompetent patients with active and incipient TB. Clin. Infect. Dis. 70, 933–936 (2019).
Cooper, A. M., Pearl, J. E., Brooks, J. V., Ehlers, S. & Orme, I. M. Expression of the nitric oxide synthase 2 gene is not essential for early control of Mycobacterium tuberculosis in the murine lung. Infect. Immun. 68, 6879–6882 (2000).
Moreira-Teixeira, L. et al. T Cell-derived IL-10 impairs host resistance to Mycobacterium tuberculosis infection. J. Immunol. 199, 613–623 (2017).
Pichugin, A. V., Yan, B. S., Sloutsky, A., Kobzik, L. & Kramnik, I. Dominant role of the sst1 locus in pathogenesis of necrotizing lung granulomas during chronic tuberculosis infection and reactivation in genetically resistant hosts. Am. J. Pathol. 174, 2190–2201 (2009).
Ji, D. X. et al. Type I interferon-driven susceptibility to Mycobacterium tuberculosis is mediated by IL-1Ra. Nat. Microbiol. 4, 2128–2135 (2019).
Esmail, H. et al. Complement pathway gene activation and rising circulating immune complexes characterize early disease in HIV-associated tuberculosis. Proc. Natl Acad. Sci. USA 115, E964–E973 (2018).
Irwin, S. M. et al. Presence of multiple lesion types with vastly different microenvironments in C3HeB/FeJ mice following aerosol infection with Mycobacterium tuberculosis. Dis. Models Mech. 8, 591–602 (2015).
Ewels, P., Magnusson, M., Lundin, S. & Kaller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Stat. Methodol. 57, 289–300 (1995).
Hadley, W. Ggplot2: Elegant Graphics for Data Analysis 2nd edn (Springer International Publishing, 2016).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015).
Chen, Z. et al. Inference of immune cell composition on the expression profiles of mouse tissue. Sci. Rep. 7, 40508 (2017).
Heng, T. S. & Painter, M. W. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008).
Yoshida, H. et al. The cis-regulatory atlas of the mouse immune system. Cell 176, 897–912 (2019).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008).
Yip, A. M. & Horvath, S. Gene network interconnectedness and the generalized topological overlap measure. BMC Bioinformatics 8, 22 (2007).
Langfelder, P., Zhang, B. & Horvath, S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 24, 719–720 (2008).
Yaari, G., Bolen, C. R., Thakar, J. & Kleinstein, S. H. Quantitative set analysis for gene expression: a method to quantify gene set differential expression including gene–gene correlations. Nucleic Acids Res. 41, e170 (2013).
Acknowledgements
The authors thank S. Hadebe (Francis Crick Institute UK, now UCT, South Africa) and J. Pitt (Medical Research Council (MRC), National Institute for Medical Research (NIMR), UK) for some of the early mouse M. tuberculosis infections and samples and sample processing leading up to the current study and K. Potempa (MRC, NIMR, UK) for early analysis of some of the microarray data leading up to the current study. The authors also thank L. Gabryšová (Francis Crick Institute UK, now Novartis, Basel, Switzerland) for her intellectual contribution to discussion of the project. The authors thank X. Wu for her help in organization of mice for TB experiments. The authors thank the Francis Crick Institute Science Technology Platforms: Biological Services for breeding and maintenance of the mice used for the early mouse M. tuberculosis infections and samples leading up to the current study; Advanced Sequencing Facility, Bioinformatics and Biostatistics Science Technology Platforms for their contribution to our sequencing processing and R. Goldstone for excellent project management of sequencing and D. Jackson for support of sequencing; and Experimental Histopathology for their excellent work in preparing lung sections for histological analyses. This study was funded by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001126), the UK MRC (FC001126) and the Wellcome Trust (FC001126); before that by the UK MRC (U117565642); and by the European Research Council (294682-TB-PATH). A.O’G., L.M-T., O.T., C.M.G., A. Singhania and E.S. were supported by the Francis Crick Institute, which receives its core funding as described above; L.M.-T., and A. Singhania, were additionally funded by the European Research Council (294682-TB-PATH). S.L.P., A.S.-B. and E.H. were funded by the Royal Veterinary College and the Francis Crick Institute. K.D.M.-B. and A. Sher were funded by the Intramural Research Program of the National Institutes of Allergy and Infectious Disease. M.S. was funded by FEDER-Fundo Europeu de Desenvolvimento Regional funds through the COMPETE 2020 – Operational Programme for Competitiveness and Internationalization (POCI), Portugal 2020 and by Portuguese funds through Fundação para a Ciência e a Tecnologia (FCT) in the framework of the project ‘Institute for Research and Innovation in Health Sciences’ (POCI-01-0145-FEDER-007274) and by FCT through Estimulo Individual ao Emprego Científico. K.L.F. and B.C. are funded by FCT PhD scholarships SFRH/BD/114405/2016 and SFRH/BD/114403/2016, respectively. P.H. and R.V. were supported by NIHR Leicester Biomedical Research Centre and the University of Leicester.
Author information
Authors and Affiliations
Contributions
A.O’G. conceived and designed the study with input from L.M.-T., M.S., C.M.G. and P.S.R. and led and coordinated the study with help from L.M.-T. A.O’G. cowrote the manuscript together with L.M.-T. O.T. performed the major part of the bioinformatics analyses with considerable input from A. Singhania, supervised by A.O’G. P.C. provided technical bioinformatics support. L.M.-T. coordinated the logistics of the study. C.M.G. isolated RNA and prepared sequencing libraries from the TB mouse models. E.S. assisted in TB mouse model experiments executed by K.L.F., J.S., B.C. and P.S.R. and designed by A.O’G., L.M.-T. and M.S. E.S. contributed to all early mouse model TB experiments executed with P.S.R. and L.M.-T., supervised by A.O’G., L.M.-T. and M.S. S.L.P., A.S.-B. and E.H. performed histopathological analysis and interpretation. K.D.M.-B. and A.Sh. provided TB samples from early mouse TB experiments leading up to the current study. M.S., K.D.M.-B. and A. Sher also provided important discussion for the project throughout and critical feedback on the manuscript. R.V. and P.H. provided clinical analysis of patients with TB and also provided critical feedback on the manuscript and important discussion for the project. All coauthors read, reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Zoltan Fehervari was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–9.
Supplementary Table 1
Human blood TB modules annotation. Annotation and number of genes of each human blood TB module (HB1–HB23) from Singhania et al.16. Module name indicates biological processes associated with the genes included in the module. Gene of interest lists a subset of genes of interest included in each module, Canonical Pathway (IPA) shows the top five canonical pathways found with IPA and Pathways (Acumenta) shows the top five pathways found with Acumenta (Singhania et al.16).
Supplementary Table 2
Human blood data in human blood TB modules, from Singhania et al.16, separated in 12 tabs depicting: London individuals raw, the raw read counts for all genes in all individual samples from the London dataset; London groups raw, the mean raw counts per group from the London dataset for all genes; London individuals norm, the normalized read counts of filtered genes in all individual samples from the London dataset; London groups norm, the mean normalized read counts per group from the London dataset for filtered genes; SA individuals raw, the raw read counts of all genes in all individual samples from the South Africa (SA) dataset; SA groups raw, the mean raw counts per group from the SA dataset for all genes; SA individuals norm, the normalized read counts of filtered genes in all individual samples of SA dataset; SA groups norm, the mean normalized read counts per group from the SA dataset for filtered genes; Leicester individuals raw, the raw read counts of all genes in all individual samples from the Leicester dataset; Leicester groups raw, the mean raw counts per group from the Leicester dataset for all genes; Leicester individuals norm, the normalized read counts of filtered genes in all individual samples from the Leicester dataset; Leicester groups norm, the mean normalized read counts per group from the Leicester dataset for filtered genes. For each gene, the corresponding human blood TB module is detailed if applicable. Normalized read counts have been processed using the variance stabilizing transformation from DESeq2 R library, and genes with low read counts have been filtered out, as described in Singhania et al.16.
Supplementary Table 3
Mouse blood data in human blood TB modules, from Singhania et al.16, separated in six tabs depicting: mouse blood individuals raw, the raw read counts of all genes in all individual samples of mouse blood RNA-seq dataset; mouse blood groups raw, the mean raw counts per group in the mouse blood RNA-seq dataset for all genes; mouse blood individuals norm, the normalized read counts of all genes in all individual samples of mouse blood RNA-seq dataset; mouse blood groups norm, the mean normalized read counts per group in the mouse blood RNA-seq dataset for all genes; Supplementary Fig. 2a mouse blood microarray, the mouse blood microarray data corresponding to Supplementary Fig. 2a; Supplementary Fig. 3a mouse blood RNA-seq, the mouse blood normalized RNA-seq data corresponding to Supplementary Fig. 3a. For each gene, the corresponding human blood TB module is detailed if applicable. Normalized read counts from RNA-seq data represent read counts processed using the variance stabilizing transformation from DESeq2 R library.
Supplementary Table 4
Mouse lung data in mouse lung disease modules, from Singhania et al.31, separated in six tabs depicting: mouse lung individuals raw, the raw read counts of all genes in all individual samples of mouse lung RNA-seq dataset; mouse lung groups raw, the mean raw counts per group in the mouse lung RNA-seq dataset for all genes; mouse lung individuals norm, the normalized read counts of all genes in all individual samples of lung blood RNA-seq dataset; mouse lung groups norm, the mean normalized read counts per group in the mouse lung RNA-seq dataset for all genes; Supplementary Fig. 2b mouse lung microarray, the mouse lung microarray data corresponding to Supplementary Fig. 2b; Supplementary Fig. 3b mouse lung RNA-seq, the mouse lung normalized RNA-seq data corresponding to Supplementary Fig. 3b. For each gene, the corresponding disease lung module is detailed if applicable. Normalized read counts from RNA-seq data represent read counts processed using the variance stabilizing transformation from DESeq2 R library.
Supplementary Table 5
Mouse lung TB modules annotation. Annotation and the number of genes for each mouse lung TB module (ML1–ML27). Module name indicates biological processes associated with the genes within the module. Gene of interest lists a subset of genes of interest included in each module and all the other columns represent an output from either IPA or Metacore.
Supplementary Table 6
Mouse lung data in mouse lung TB modules, separated in four tabs depicting: mouse lung individuals raw, the raw read counts of all genes in all individual samples of mouse lung RNA-seq dataset; Mouse lung groups raw, the mean raw counts per group in the mouse lung RNA-seq dataset for all genes; Mouse lung individuals norm, the normalized read counts of all genes in all samples of mouse lung RNA-seq dataset; Mouse lung groups norm, the mean normalized read counts per group in the mouse lung RNA-seq dataset for all genes. For each gene, the corresponding TB lung module is detailed if applicable. Normalized read counts from RNA-seq data represent read counts processed using the variance stabilizing transformation from DESeq2 R library.
Supplementary Table 7
X-ray scores for human TB cohort from Leicester. X-ray classification (CXR classification) for each patient evaluated from the Leicester cohort and their associated disease group (TB subgroup).
Supplementary Table 8
Mouse to human gene mapping. The table lists the correspondence between mouse Ensembl gene ID and human Ensembl gene ID (ortholog genes), found using biomaRt package in R.
Rights and permissions
About this article
Cite this article
Moreira-Teixeira, L., Tabone, O., Graham, C.M. et al. Mouse transcriptome reveals potential signatures of protection and pathogenesis in human tuberculosis. Nat Immunol 21, 464–476 (2020). https://doi.org/10.1038/s41590-020-0610-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-020-0610-z
This article is cited by
-
Autophagy promotes efficient T cell responses to restrict high-dose Mycobacterium tuberculosis infection in mice
Nature Microbiology (2024)
-
Catheter-associated Mycobacterium intracellulare biofilm infection in C3HeB/FeJ mice
Scientific Reports (2023)
-
Host-directed immunotherapy of viral and bacterial infections: past, present and future
Nature Reviews Immunology (2023)
-
BACH1 promotes tissue necrosis and Mycobacterium tuberculosis susceptibility
Nature Microbiology (2023)
-
Lung gene expression signatures suggest pathogenic links and molecular markers for pulmonary tuberculosis, adenocarcinoma and sarcoidosis
Communications Biology (2020)