Abstract
Chronic cytomegalovirus (CMV) infection leads to long-term maintenance of extraordinarily large CMV-specific T cell populations. The magnitude of this so-called ‘memory inflation’ is thought to mainly depend on antigenic stimulation during the chronic phase of infection. However, by mapping the long-term development of CD8+ T cell families derived from single naive precursors, we find that fate decisions made during the acute phase of murine CMV infection can alter the level of memory inflation by more than 1,000-fold. Counterintuitively, a T cell family’s capacity for memory inflation is not determined by its initial expansion. Instead, those rare T cell families that dominate the chronic phase of infection show an early transcriptomic signature akin to that of established T central memory cells. Accordingly, a T cell family’s long-term dominance is best predicted by its early content of T central memory precursors, which later serve as a stem-cell-like source for memory inflation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
RNA sequencing data generated for this study have been deposited in the Gene Expression Omnibus under accession number GSE157644. All other data generated or analyzed during this study are included in the published article (and its supplementary information files) or are available from the corresponding author upon reasonable request. Source data are provided with this paper.
Code availability
No custom code or algorithms were used in this study.
References
Karrer, U. et al. Memory inflation: continuous accumulation of antiviral CD8+ T cells over time. J. Immunol. 170, 2022–2029 (2003).
Sylwester, A. W. et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 202, 673–685 (2005).
Simon, C. O. et al. CD8 T cells control cytomegalovirus latency by epitope-specific sensing of transcriptional reactivation. J. Virol. 80, 10436–10456 (2006).
Dekhtiarenko, I. et al. Peptide processing is critical for T-cell memory inflation and may be optimized to improve immune protection by CMV-based vaccine vectors. PLoS Pathog. 12, e1006072 (2016).
Snyder, C. M. et al. Memory inflation during chronic viral infection is maintained by continuous production of short-lived, functional T cells. Immunity 29, 650–659 (2008).
Klenerman, P. & Oxenius, A. T cell responses to cytomegalovirus. Nat. Rev. Immunol. 16, 367–377 (2016).
Karrer, U. et al. Expansion of protective CD8+ T-cell responses driven by recombinant cytomegaloviruses. J. Virol. 78, 2255–2264 (2004).
Hansen, S. G. et al. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat. Med. 15, 293–299 (2009).
Hansen, S. G. et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473, 523–527 (2011).
Hansen, S. G. et al. Immune clearance of highly pathogenic SIV infection. Nature 502, 100–104 (2013).
Hansen, S. G. et al. Prevention of tuberculosis in rhesus macaques by a cytomegalovirus-based vaccine. Nat. Med. 24, 130–143 (2018).
Weekes, M. P., Wills, M. R., Mynard, K., Carmichael, A. J. & Sissons, J. G. The memory cytotoxic T-lymphocyte (CTL) response to human cytomegalovirus infection contains individual peptide-specific CTL clones that have undergone extensive expansion in vivo. J. Virol. 73, 2099–2108 (1999).
Khan, N., Cobbold, M., Keenan, R. & Moss, P. A. H. Comparative analysis of CD8+ T cell responses against human cytomegalovirus proteins pp65 and immediate early 1 shows similarities in precursor frequency, oligoclonality, and phenotype. J. Infect. Dis. 185, 1025–1034 (2002).
McHeyzer-Williams, M. G. & Davis, M. M. Antigen-specific development of primary and memory T cells in vivo. Science 268, 106–111 (1995).
Lin, M. Y. & Welsh, R. M. Stability and diversity of T cell receptor repertoire usage during lymphocytic choriomeningitis virus infection of mice. J. Exp. Med. 188, 1993–2005 (1998).
Busch, D. H. & Pamer, E. G. T cell affinity maturation by selective expansion during infection. J. Exp. Med. 189, 701–710 (1999).
Zehn, D., Lee, S. Y. & Bevan, M. J. Complete but curtailed T-cell response to very low-affinity antigen. Nature 458, 211–214 (2009).
Williams, M. A., Ravkov, E. V. & Bevan, M. J. Rapid culling of the CD4+ T cell repertoire in the transition from effector to memory. Immunity 28, 533–545 (2008).
Kim, C., Wilson, T., Fischer, K. F. & Williams, M. A. Sustained interactions between T cell receptors and antigens promote the differentiation of CD4+ memory T cells. Immunity 39, 508–520 (2013).
Schober, K. et al. Reverse TCR repertoire evolution toward dominant low-affinity clones during chronic CMV infection. Nat. Immunol. 21, 434–441 (2020).
Day, E. K. et al. Rapid CD8+ T cell repertoire focusing and selection of high-affinity clones into memory following primary infection with a persistent human virus: human cytomegalovirus. J. Immunol. 179, 3203–3213 (2007).
Trautmann, L. et al. Selection of T cell clones expressing high-affinity public TCRs within human cytomegalovirus-specific CD8 T cell responses. J. Immunol. 175, 6123–6132 (2005).
Buchholz, V. R. et al. Disparate individual fates compose robust CD8+ T cell immunity. Science 340, 630–635 (2013).
Gerlach, C. et al. Heterogeneous differentiation patterns of individual CD8+ T cells. Science 340, 635–639 (2013).
Cho, Y.-L. et al. TCR signal quality modulates fate decisions of single CD4+ T cells in a probabilistic manner. Cell Rep. 20, 806–818 (2017).
Buchholz, V. R., Schumacher, T. N. M. & Busch, D. H. T cell fate at the single-cell level. Annu. Rev. Immunol. 34, 65–92 (2016).
La Manno, G. et al. RNA velocity of single cells. Nature 560, 494–498 (2018).
Plumlee, C. R., Sheridan, B. S., Cicek, B. B. & Lefrançois, L. Environmental cues dictate the fate of individual CD8+ T cells responding to infection. Immunity 39, 347–356 (2013).
Tubo, N. J. et al. Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell 153, 785–796 (2013).
Grassmann, S. et al. Distinct surface expression of activating receptor Ly49H drives differential expansion of NK cell clones upon murine cytomegalovirus infection. Immunity 50, 1391–1400.e4 (2019).
Dekhtiarenko, I., Jarvis, M. A., Ruzsics, Z. & Cicin-Sain, L. The context of gene expression defines the immunodominance hierarchy of cytomegalovirus antigens. J. Immunol. 190, 3399–3409 (2013).
Graef, P. et al. Serial transfer of single-cell-derived immunocompetence reveals stemness of CD8+ central memory T cells. Immunity 41, 116–126 (2014).
Flossdorf, M., Rössler, J., Buchholz, V. R., Busch, D. H. & Höfer, T. CD8+ T cell diversification by asymmetric cell division. Nat. Immunol. 16, 891–893 (2015).
Kretschmer, L. et al. Differential expansion of T central memory precursor and effector subsets is regulated by division speed. Nat. Commun. 11, 113 (2020).
Jeannet, G. et al. Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc. Natl Acad. Sci. USA 107, 9777–9782 (2010).
Zhou, X. et al. Differentiation and persistence of memory CD8+ T cells depend on T cell factor 1. Immunity 33, 229–240 (2010).
Shrestha, S. et al. Tsc1 promotes the differentiation of memory CD8+ T cells via orchestrating the transcriptional and metabolic programs. Proc. Natl Acad. Sci. USA 111, 14858–14863 (2014).
Miao, T. et al. Egr2 and 3 control adaptive immune responses by temporally uncoupling expansion from T cell differentiation. J. Exp. Med. https://doi.org/10.1084/jem.20160553 (2017).
Jeng, M. Y. et al. Metabolic reprogramming of human CD8+ memory T cells through loss of SIRT1. J. Exp. Med. 215, 51–62 (2018).
Byrne, S. M. et al. Cathepsin B controls the persistence of memory CD8+ T lymphocytes. J. Immunol. 189, 1133–1143 (2012).
Yeh, J.-H., Sidhu, S. S. & Chan, A. C. Regulation of a late phase of T cell polarity and effector functions by Crtam. Cell 132, 846–859 (2008).
Im, S. J. et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421 (2016).
van der Windt, G. J. W. et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78 (2012).
Gulati, G. S. et al. Single-cell transcriptional diversity is a hallmark of developmental potential. Science 367, 405–411 (2020).
Joshi, N. S. et al. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity 27, 281–295 (2007).
Sarkar, S. et al. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med. 205, 625–640 (2008).
Obar, J. J. & Lefrançois, L. Early signals during CD8 T cell priming regulate the generation of central memory cells. J. Immunol. 185, 263–272 (2010).
Lin, W.-H. W. et al. CD8+ T lymphocyte self-renewal during effector cell determination. Cell Rep. 17, 1773–1782 (2016).
Welten, S. P. M. et al. Tcf1+ cells are required to maintain the inflationary T cell pool upon MCMV infection. Nat. Commun. 11, 2295 (2020).
Davenport, M. P., Fazou, C., McMichael, A. J. & Callan, M. F. C. Clonal selection, clonal senescence, and clonal succession: the evolution of the T cell response to infection with a persistent virus. J. Immunol. 168, 3309–3317 (2002).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Young, M. D., Wakefield, M. J., Smyth, G. K. & Oshlack, A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14 (2010).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
McCarthy, D. J., Campbell, K. R. & Lun, A. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186 (2017).
DeTomaso, D. & Yosef, N. FastProject: a tool for low-dimensional analysis of single-cell RNA-Seq data. BMC Bioinformatics 17, 315 (2016).
Bergen, V., Lange, M., Peidli, S., Wolf, F. A. & Theis, F. J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. https://doi.org/10.1038/s41587-020-0591-3 (2020).
Haghverdi, L., Büttner, M., Wolf, F. A., Buettner, F. & Theis, F. J. Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 13, 845–848 (2016).
Acknowledgements
We thank thank J. Sun and N. Adams as well as all members of the Buchholz laboratory for discussion and support. This work was supported by the Else Kröner-Fresenius-Stiftung – EKFS 2019_A91 (‘Dissecting the induction of inflationary CD8+ T cell memory on the single-cell level’) to V.R.B., the DFG – SFB 1054 (project no. 210592381) to V.R.B. and D.H.B., the TUM Seed Funds (SCIMAP) to V.R.B., the BMBF project Quan-T-cell (e:Med initiative on Systems Medicine, FKZ 01ZX1505) to M.F., the BMBF project TIDY (Computational Life Sciences, FKZ 031L0170A) to T.H., the Helmholtz association project PIE-008 to L.C.S. and the DFG – TRR 179 (project number 031L0170A) to M.S.
Author information
Authors and Affiliations
Contributions
S.G. and V.R.B. designed the study. S.G., L.M., R.R., S.F., I.H., J.L., L.K. and L.O.P. performed experiments. S.G., L.M., J.M., A.K., A.J. and M.F. performed data analysis. Q.Z., A.J. and T.H. provided sample processing and RNA sequencing. M.Z.C. and L.C.-S. generated virus stocks. M.S. provided cell sorting. K.S. and D.H.B. contributed to study design and critically read the manuscript. S.G., L.M. and V.R.B. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Peer reviewer reports are available. Zoltan Fehervari was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
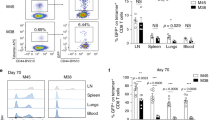
Extended Data Fig. 1 Virus schematic, gating strategy and tracking of T cell families until day 412 post infection.
a, Schematic of MCMV-ie2-SIINFEKL virus. The SIINFEKL peptide sequence was inserted at the 3’ end of the ie2 gene. b, Gating strategy for single-cell fate mapping. Following doublet and dead-cell exclusion, transferred cells are identified by expression of congenic markers. Further gating depends on the method used for single-cell transfer (exemplified in Figs. 2b and 4b, respectively). T cell families are characterized phenotypically using antibodies directed against CD27 and CD62L. c, Longitudinal quantification of T cell family sizes in blood, measured as percent of living leukocytes, for a cohort of T cell families from Fig. 1e traced until day 412 p.i. n = 15 T cell families.
Extended Data Fig. 2 Gene ontology term enrichment analysis of early gene signature in long-term dominators (sc-derived RNA-seq, day 8 p.i.).
a–c, Gene ontology (GO) term enrichment analysis of early gene signature in long-term dominating T cell families acquired via sc-derived RNA-seq at day 8 p.i. a, Top 10 GO terms that were underrepresented in long-term dominators (n = 2) compared to all remaining T cell families (n = 14). b,c, Top 10 GO terms that were significantly underrepresented (b) or overrepresented (c) in long-term dominators (n = 2) compared to acutely dominating T cell families (n = 2). d, GSEA analysis of genes differentially expressed in long-term dominating vs. acutely dominating T cell families associated with cellular senescence (GO:0090398). Genes up- (red) and down-regulated (blue) in long-term dominators vs. acutely dominating T cell families. P values were determined based on the default method in the goseq (a–c) and fgsea packages (d). See methods for details.
Extended Data Fig. 3 Expression of genes enriched in TCM, TEM and TE clusters (scRNA-seq, day 60 p.i.).
a,b, Analysis of scRNA-seq data derived from an established inflationary T cell population in spleen at day 60 p.i. a, Dimensional reduction using Uniform Manifold Approximation and Projection (UMAP) showing Leiden clusters and expression of genes enriched in the corresponding TCM, TEM and TE clusters. b, Cell cycle scores for S and G2M-phase projected onto UMAP embedding. These scores indicate both a weak proliferative activity and a uniform distribution of scores across different Leiden clusters. Scores were computed based on lists for S- and G2M-phase associated genes using the software package SCANPY.
Extended Data Fig. 4 Retrogenic color-barcoding and single-cell transfer.
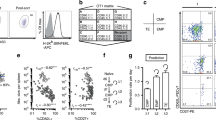
a, Schematic of the generation of retrogenic color-barcoded OT-1 T cells: Bone marrow of CD45.1+ OT-1 Rag1–/– mice was harvested and enriched for hematopoietic stem cells (HSCs) by flow cytometric sorting for Sca1+ cells. These were transduced retrovirally with different combinations of fluorescent proteins (GFP, YFP, BFP, CFP and T-sapphire) and transplanted into sublethally irradiated C57BL/6 mice. More than 6 weeks after HSC engraftment, naive color-barcoded OT-1 T cells were harvested for experiments from peripheral blood of retrogenic chimeras. b, Exemplary gating strategy for identification of color-barcoded naive OT-1 T-cells. c, Sort and transfer of single naive color-barcoded OT-1 T cells (see methods section for details).
Extended Data Fig. 5 Temporal dynamics of single-cell-derived memory inflation correlated to early abundance of TEs, EMPs and CMPs.
a, Scatter plots comparing the amount of TEs (top row), EMPs (middle row) and CMPs (bottom row) in each T cell family on day 8 p.i. to the total size of the respective T cell families at various time points between day 8 and day 140 p.i. The amount of CMPs, EMPs and TEs and the total size of T cell families is measured as % of living leukocytes. Top-10 long-term dominating T cell families are marked (blue circles). P values from left to right: TE: p < 0.0001, p < 0.0001, p = 0.009, p = 0.95, p = 0.91, p = 0.97. EMP: p < 0.0001, p < 0.0001, p = 0.003, p = 0.88, p = 0.30, p = 0.48. CMP: p = 0.001, p = 0.001, p = 0.007, p = 0.0018, p = 0.0008, p = 0.0007. b, Scatter plots comparing the relative amount of TEs (top row), EMPs (middle row) and CMPs (bottom row) in each T cell family on day 8 p.i. to the total size of the respective T cell families at various time points between day 8 and day 140 p.i. The relative amount of CMPs, EMPs and TEs is measured as % of the corresponding T cell family’s overall size. Top-10 long-term dominating T cell families are marked (blue circles). P values from left to right: TE: p = 0.99, p = 0.07, p = 0.15, p = 0.13, p = 0.006, p = 0.002. EMP: p = 0.59, p = 0.73, p = 0.90, p = 0.82, p = 0.39, p = 0.10. CMP: p < 0.0001, p = 0.58, p = 0.57, p = 0.42, p = 0.0008, p = 0.0004. ‘r’ indicates Spearman’s correlation coefficient. Significances were measured using a two-sided Mann-Whitney test. * p < 0.05, ** p <0.01, *** p <0.001, **** p < 0.0001. n = 104 T cell families (zero values are not shown and not used for determining Spearman’s correlation coefficients and respective p values).
Extended Data Fig. 6 CMPs constitute only a minute fraction of MPECs and are enriched for non-cytolytic T cells.
a–d, Adoptive transfer of 100 congenically labelled OT-1 T cells followed by immunization with MCMV-ie2-SIINFEKL. a,b, Analysis of CD27, CD127, KLRG-1 and CD62L expression in expanded T cell populations (n = 6) at day 8 p.i. in spleen. a, Percent of CD62L+CD27+ CMPs among all responding T cells or among KLRG-1–CD27+ MPECs b, Percent of CD62L+CD27+ CMPs among all responding T cells or KLRG-1–CD127hi MPECs or KLRG-1+CD127lo SLECs. c,d, Analysis of Gzmb expression in T cell subsets defined as in (a) measured in expanded T cell populations (n = 5) at day 8 p.i. c, Percent of Gzmb– cells among MPECs or SLECs (d) Percent of Gzmb– cells among CMPs, EMPs or TEs. e,f, Adoptive transfer of 100 congenically labelled OT-1 T cells followed by immunization with 5000 cfu L.m.-OVA. Analysis of Gzmb expression in T cell subsets defined as in (a) in expanded T cell populations (n = 4) at day 8 p.i. e, Percent of Gzmb– cells among MPECs or SLECs (f) Percent of Gzmb– cells among CMPs, EMPs or TEs. Stacked bars in 6a–f show mean, error bars indicate SD. g,h, Further analysis of experiments described in Fig. 4: Scatter plots comparing the amount of CD27+ Memory precursors (MPs) (g) or the percentage of MPs in each T cell family (h) on day 8 p.i. to the total size of the respective T cell families at various time points between day 8 and day 140 p.i. The amount of MPs and the total size of T cell families is measured as % of living leukocytes. The relative amount of MPs is measured as % of the corresponding T cell family’s overall size. Top-10 long-term dominating T cell families are marked (blue circles). P values from left to right: (g): TE: p < 0.0001, p < 0.0001, p = 0.008, p = 0.69, p = 0.16, p = 0.30. (h): p = 0.14, p = 0.21, p = 0.27, p = 0.19, p = 0.01, p = 0.002. ‘r’ indicates Spearman’s correlation coefficient. Significances were measured using a two-sided Mann-Whitney test. * p < 0.05, ** p <0.01, *** p <0.001, **** p < 0.0001. n = 104 T cell families (zero values are not shown and not used for determining Spearman’s correlation coefficients and respective p values).
Supplementary information
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data and lists of differentially expressed genes.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Lists of differentially expressed genes.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
About this article
Cite this article
Grassmann, S., Mihatsch, L., Mir, J. et al. Early emergence of T central memory precursors programs clonal dominance during chronic viral infection. Nat Immunol 21, 1563–1573 (2020). https://doi.org/10.1038/s41590-020-00807-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-020-00807-y
This article is cited by
-
B cells orchestrate tolerance to the neuromyelitis optica autoantigen AQP4
Nature (2024)
-
Granzyme B + CD8 + T cells with terminal differentiated effector signature determine multiple sclerosis progression
Journal of Neuroinflammation (2023)
-
Unbiased chemokine receptor screening reveals similar efficacy of lymph node- and tumor-targeted T cell immunotherapy
Cancer Immunology, Immunotherapy (2023)
-
Picking up speed: cell cycle regulation during effector CD8+ T cell differentiation
Medical Microbiology and Immunology (2023)
-
SARS-CoV-2 infection establishes a stable and age-independent CD8+ T cell response against a dominant nucleocapsid epitope using restricted T cell receptors
Nature Communications (2023)