Abstract
Here we identify a group 2 innate lymphoid cell (ILC2) subpopulation that can convert into interleukin-17 (IL-17)-producing NKp44− ILC3-like cells. c-Kit and CCR6 define this ILC2 subpopulation that exhibits ILC3 features, including RORγt, enabling the conversion into IL-17-producing cells in response to IL-1β and IL-23. We also report a role for transforming growth factor-β in promoting the conversion of c-Kit− ILC2s into RORγt-expressing cells by inducing the upregulation of IL23R, CCR6 and KIT messenger RNA in these cells. This switch was dependent on RORγt and the downregulation of GATA-3. IL-4 was able to reverse this event, supporting a role for this cytokine in maintaining ILC2 identity. Notably, this plasticity has physiological relevance because a subset of RORγt+ ILC2s express the skin-homing receptor CCR10, and the frequencies of IL-17-producing ILC3s are increased at the expense of ILC2s within the lesional skin of patients with psoriasis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Sequence data that support the findings of this study have been deposited in the Gene Expression Omnibus with accession numbers GSE124474, GSE129238 and GSE114396 for bulk RNAseq data (Fig. 3) for ILC subsets, microarray data of human ILC2 subsets cultured under IL-17-polarizing conditions (Fig. 4) and scRNAseq data of human ILC2s (Fig. 5), respectively. The data that support the findings from these studies are available from the corresponding authors upon request.
Change history
13 November 2019
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Artis, D. & Spits, H. The biology of innate lymphoid cells. Nature 517, 293–301 (2015).
Spits, H. et al. Innate lymphoid cells—a proposal for a uniform nomenclature. Nat. Rev. Immunol. 13, 145–149 (2013).
Bernink, J. H. et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat. Immunol. 14, 221–229 (2013).
Bal, S. M. et al. IL-1beta, IL-4 and IL-12 control the fate of group 2 innate lymphoid cells in human airway inflammation in the lungs. Nat. Immunol. 17, 636–645 (2016).
Ohne, Y. et al. IL-1 is a critical regulator of group 2 innate lymphoid cell function and plasticity. Nat. Immunol. 17, 646–655 (2016).
Papp, K. A. et al. Risankizumab versus ustekinumab for moderate-to-severe plaque psoriasis. N. Engl. J. Med. 376, 1551–1560 (2017).
Hedrick, M. N. et al. CCR6 is required for IL-23-induced psoriasis-like inflammation in mice. J. Clin. Invest. 119, 2317–2329 (2009).
Pantelyushin, S. et al. Rorgammat+innate lymphocytes and gammadelta T cells initiate psoriasiform plaque formation in mice. J. Clin. Invest. 122, 2252–2256 (2012).
Bjorklund, A. K. et al. The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. Nat. Immunol. 17, 451–460 (2016).
Munneke, J. M. et al. Activated innate lymphoid cells are associated with a reduced susceptibility to graft-versus-host disease. Blood 124, 812–821 (2014).
Teunissen, M. B. et al. Composition of innate lymphoid cell subsets in the human skin: enrichment of NCR(+) ILC3 in lesional skin and blood of psoriasis patients. J. Invest. Dermatol. 134, 2351–2360 (2014).
Villanova, F. et al. Characterization of innate lymphoid cells in human skin and blood demonstrates increase of NKp44+ILC3 in psoriasis. J. Invest. Dermatol. 134, 984–991 (2014).
Huang, Y. et al. IL-25-responsive, lineage-negative KLRG1(hi) cells are multipotential ‘inflammatory’ type 2 innate lymphoid cells. Nat. Immunol. 16, 161–169 (2015).
Ricardo-Gonzalez, R. R. et al. Tissue signals imprint ILC2 identity with anticipatory function. Nat. Immunol. 19, 1093–1099 (2018).
Mjosberg, J. et al. The transcription factor GATA3 is essential for the function of human type 2 innate lymphoid cells. Immunity 37, 649–659 (2012).
Mjosberg, J. M. et al. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat. Immunol. 12, 1055–1062 (2011).
Lim, A. I. et al. Systemic human ILC precursors provide a substrate for tissue ILC differentiation. Cell 168, 1086–1100.e1010 (2017).
Miossec, P., Korn, T. & Kuchroo, V. K. Interleukin-17 and type 17 helper T cells. N. Engl. J. Med. 361, 888–898 (2009).
Ivanov, I. I. et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+T helper cells. Cell 126, 1121–1133 (2006).
Okada, S. et al. IMMUNODEFICIENCIES. impairment of immunity to candida and mycobacterium in humans with bi-allelic RORC mutations. Science 349, 606–613 (2015).
Hoyler, T. et al. The transcription factor GATA-3 controls cell fate and maintenance of Type 2 innate lymphoid cells. Immunity 37, 634–648 (2012).
Bernink, J. H. et al. Interleukin-12 and -23 control plasticity of CD127(+) group 1 and group 3 innate lymphoid cells in the intestinal lamina propria. Immunity 43, 146–160 (2015).
Silver, J. S. et al. Inflammatory triggers associated with exacerbations of COPD orchestrate plasticity of group 2 innate lymphoid cells in the lungs. Nat. Immunol. 17, 1005 (2016).
Diefenbach, A., Colonna, M. & Koyasu, S. Development, differentiation, and diversity of innate lymphoid cells. Immunity 41, 354–365 (2014).
Turner, J. E. et al. IL-9-mediated survival of type 2 innate lymphoid cells promotes damage control in helminth-induced lung inflammation. J. Exp. Med. 210, 2951–2965 (2013).
Nussbaum, J. C. et al. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature 502, 245–248 (2013).
Robinette, M. L. et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 16, 306–317 (2015).
Haque, A., Engel, J., Teichmann, S. A. & Lonnberg, T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 9, 75 (2017).
McCully, M. L. & Moser, B. The human cutaneous chemokine system. Front. Immunol. 2, 33 (2011).
Ullrich, E. et al. BATF-dependent IL-7RhiGM-CSF+T cells control intestinal graft-versus-host disease. J. Clin. Invest. 128, 916–930 (2018).
Hughes, T. et al. Interleukin-1beta selectively expands and sustains interleukin-22+immature human natural killer cells in secondary lymphoid tissue. Immunity 32, 803–814 (2010).
Crellin, N. K. et al. Regulation of cytokine secretion in human CD127(+) LTi-like innate lymphoid cells by Toll-like receptor 2. Immunity 33, 752–764 (2010).
Cella, M., Otero, K. & Colonna, M. Expansion of human NK-22 cells with IL-7, IL-2, and IL-1beta reveals intrinsic functional plasticity. Proc. Natl Acad. Sci. USA 107, 10961–10966 (2010).
Lim, A. I. et al. IL-12 drives functional plasticity of human group 2 innate lymphoid cells. J. Exp. Med. 213, 569–583 (2016).
Manel, N., Unutmaz, D. & Littman, D. R. The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORgammat. Nat. Immunol. 9, 641–649 (2008).
Trifari, S., Kaplan, C. D., Tran, E. H., Crellin, N. K. & Spits, H. Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)-17, T(H)1 and T(H)2 cells. Nat. Immunol. 10, 864–871 (2009).
Nestle, F. O., Kaplan, D. H. & Barker, J. Psoriasis. N. Engl. J. Med. 361, 496–509 (2009).
Boehncke, W. H. & Schon, M. P. Psoriasis. Lancet 386, 983–994 (2015).
Huang, Y. et al. S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science 359, 114–119 (2018).
Bielecki, P. et al. Skin inflammation driven by differentiation of quiescent tissue-resident ILCs into a spectrum of pathogenic effectors. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/461228v2 (2018).
Golebski, K. et al. IL-1β, IL-23, and TGF-β drive plasticity of human ILC2s towards IL-17-producing ILCs in nasal inflammation. Nat. Commun. 10, 2162 (2019).
Wang, Y. et al. Discovery of novel N-(5-(arylcarbonyl)thiazol-2-yl)amides and N-(5-(arylcarbonyl)thiophen-2-yl)amides as potent RORgammat inhibitors. Bioorganic Med. Chem. 22, 692–702 (2014).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Wallrapp, A. et al. The neuropeptide NMU amplifies ILC2-driven allergic lung inflammation. Nature 549, 351–356 (2017).
Acknowledgements
We thank B. Hooibrink for help with flow cytometry. We thank P.I. Spuls and staff at the Department of Dermatology, Academic Medical Center, Amsterdam, the Netherlands, for providing tissue samples of patients with psoriasis and W.G. van Selms and staff at the Department of Plastic Surgery, St. Lucas Andreas Hospital, Amsterdam, the Netherlands, for healthy human skin tissue samples. We thank the MedImmune Flow Cytometry core, C. Groves, G. Rosignoli, R. Rayanki and R. Grady for the support with flow cytometry and cell sorting; MedImmune Phamacogenomics group, R. Raja, R. Halpin and Y. Lee for the support of genomics experiments; I. Nikaido for the advice on processing RNAseq samples; A. Berlin, A. Copenhaver, R. Dagher, M. Yang, O. Wyatt, H. Tompkins and A. Gonzales for laboratory support; Y.J. Liu for the initial support for this work; J.S. Silver, J. Kearley and S. Rieder for the critical discussion; the LAR staff for maintaining the experimental animals; and S. Farshadi for helping figure preparation.
Author information
Authors and Affiliations
Contributions
J.H.B., Y.O., M.B.M.T. and X.R.R. designed the study, performed experiments, analyzed the data and wrote the manuscript. J. Wang, J. Wu and Y.S. performed single-cell and bulk RNAseq experiments and analyses. M.A.d.R. provided psoriatic tissue. L.K., C.G., S.v.T. and I.R. performed experiments and analyzed data. R.V. and J.K. analyzed microarray data. H.S. and A.A.H. designed the study, analyzed the data and wrote the manuscript. H.S. is supported by an advanced grant of the European Research Council ERC-2013-ADG number 341038.
Corresponding authors
Ethics declarations
Competing interests
Y.O., X.R.R., J.Wang, Y.S. and A.A.H. are employed by and shareholders of AstraZeneca, and J.Wu and C.G. are employed by Novartis. H.S. is employed part time by AIMM Therapeutics.
Additional information
Peer review information Zoltan Fehervari was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 IL-17-producing ILCs activated by C. albicans originate from ILC2s.
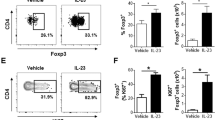
(a) Flow cytometory analysis of intracellular IL-17 levels following restimulation with PMA and ionomycin of Lin−CD127+CD161+ total ILCs (gating as in Fig. 1a) from healthy skin that have been stimulated for 6 days with C. albicans hyphae (left panel), in the presence of autologous irradiated dermal cells (middle panel) or in the presence of both irradiated dermal cells and C. albicans hyphae (right panel). Shown data is representative of 3 independent experiments. (b) Phenotypical characterization of hyphen-stimulated IL-17-secreting ILCs based on the expression of CRTH2 and c-Kit. Data shown (left) are representative of 3 independent experiments with 1 to 2 donors each that is quantified in bar diagram (right). Numbers adjacent to outlined areas indicate percent IL-17A+ cells and in quadrants indicate percent cells throughout. (c). Dermal ILC subsets were sorted as described in Fig. 1a and analysed for IL17A gene expression. Combined data of 3 experiments with independent donors. Relative units (AU) relative to the expression of β-actin. Error bars represent SEM. (d) Flow cytometry phenotyping of surface receptor expression in ILC populations from healthy skin gated as in Fig. 1a. Data are representative of 3 independent experiments with independent donors. (e) Flow cytometry gating strategy for ILC subsets from human peripheral blood, based on the expression of CD127, CD161, lineage markers (Lin) (CD1a, CD3, CD14, CD16, CD19, CD34, CD94, CD123, BDCA2, FcεR1a, TCRαβ, TCRγδ), CRTH2 and c-Kit. Numbers in gates or quadrants indicate percent cell in each. Data are representative of at least 3 independent experiments. (f) Expression of CD62L and CD69 in peripheral blood ILC2s of healthy individuals; light gray-shading, isotype-matched control antibody. Data are representative of at least 3 independent experiments. (g) Isolated ILC2s from dermis and peripheral blood from 2 healthy donors were analysed for IL5 and IL17A mRNA expression by qPCR. Relative units (RU) relative to the expression of β-actin. Bar diagrams show mean ± SEM. (h) IL-1β, IL-23 and TGF-β as detected by ELISA in the supernatant of C. albicans hyphae-stimulated irradiated dermal cells or irradiated dermal cells alone. Accumulative data of 3 experiments with one donor each. Diagrams show mean ± SEM. (i) Flow cytometry analysis of frequencies of ILC2-derived IL-17A+ cells (as in Fig. 1a) supplemented with anti-IL-1β, anti-TGF-β, anti-IL-23 (all 5 µg/ml). Bar graphs is quantification of 3 independent experiments with 1 donor each. 1-way ANOVA followed by Tukey’s test; * P < 0.05. Error bars represent SEM. Numbers adjacent to outlined areas indicate percent IL-17A+ cells. (j) Flow cytometry analysis of intracellular IL-17, IL-22, IFN-γ and IL-5 of blood-derived CRTH2-c-Kit+ ILCs following stimulation with IL-1β, IL-23 and TGF-β and restimulation with PMA and ionomycin. Data are representative of at least 3 independent donors. Number in quadrants and gates indicate percentage of cells in each. (k) Flow cytometry analyzing the expression of CRTH2, c-Kit and NKp44 in ILC2s cultured in the presence of IL-2 only (dotted line) or IL-2, IL-1β, IL-23 and TGF-β (black line) for 7 days; filled grey is isotype-matched control antibody. Data are representative of 4 experiments with independent donors. (l) Flow cytometry analysis of intracellular IL-17A and IL-5 in blood ILC2s cultured with indicated cytokines for 7 days followed by stimulation with PMA and ionomycin. The plots are representative of 3 experiments with independent donors (right). The bar diagram is quantifying frequencies of IL-17+ cells (n = 3, independent donors). Number in quadrants and gates indicate percentage of cells in each. 1-way ANOVA followed by Tukey’s test; * P < 0.05; ** P < 0.01; Error bars represent SEM. (m) Flow cytometry analysis of intracellular IL-17A, IL-13 and IL-5 in blood ILC2s cultured with IL-1β, IL-23 and TGF-β ± IL-33 for 7 days followed by stimulation with PMA and ionomycin. The plots are representative of 3 experiments with independent donors (right). The bar diagram is quantifying frequencies of IL-17+, IL-13+, IL-5+ cells (n = 3, independent donors). Number in quadrants and gates indicate percentage of cells in each. 1-way ANOVA followed by Tukey’s test; ** P < 0.01; *** P < 0.001; NS not significant. Error bars represent SEM.
Supplementary Figure 2 IL-17 production in ILC2s is controlled by the reciprocal regulation of RORγt and GATA-3 activity, and blocked by IL-4.
(a) RORγt and RORα inhibition of promoter activity in transfected cells with RORγ/GAL4 and RORα/GAL4 reporter systems in the presence of various concentration of a ROR γt inhibitor (compound 8h) as judged by luciferase reporter activity. IC50; half minimal inhibitory concentration. RLU; Relative light units. SD; standard deviation. Concentration-response curves were generated and IC50 values were calculated by nonlinear fitting using the GraphPad Prism software package. Representative example of concentration-response curves from two experiments with duplicate readings is shown. (b) RORC mRNA of Th cells under TH17 polarization conditions in the presence of various concentrations of the RORγt inhibitor as quantified by RT-qPCR. Individual data and mean ± SD are depicted. Results are representative of three independent experiments with triplicate readings. (c, d) IL-17 secretion by TH17 (c) and Tc17 cells (d) in the presence of increased concentrations of the RORγt inhibitor examined by ELISA. Concentration-response curves were generated and IC50 values were calculated by nonlinear fitting using the GraphPad Prism software package. Representative examples of concentration-response curves from at least three experiments with triplicate readings are shown. (e) IL17A, IL17F and CCR6 expression in purified human CD4+ T-cells polarized towards Th17 cells and treated at the beginning of the cell culture with various concentrations of the RORγt inhibitor or with DMSO for 72 hours examined by RT-qPCR. Gene expression was normalized to GUSB levels and expressed as arbitrary units. Individual data and mean ± SD are shown. Results are representative of three independent experiments with triplicate readings. (f) IL-17A amount quantified by ELISA in the supernatant of blood-derived ILC2s and ILC3s transduced with a retroviral vector encoding GATA3 wild-type (wt) or an empty vector, sorted as transduced GFP+ cells on day 3 and cultured with either IL-1β, IL-23 and TGF-β and (white) or IL-1β and IL-4 (black) for 7 days. (g) RORC expression following stimulation with IL-1β, IL-23 and TGF-β for 7 days of blood-derived ILC2s that are transduced with a retroviral vector encoding either an empty vector or a GATA3 wt vector. (h) GATA3 expression following stimulation with IL-1β, IL-23 and TGF-β in the presence or absence of a RORγt inhibitor, and restimulation with PMA and ionomycin of blood-derived ILC2s. Arbitrary units (AU) relative to the expression of ACTB. NS, not significant. Bar graphs is quantification of 3 independent experiments with 1 donor each. (i) Flow cytometry analysis of IL-5 and IL-17 production from ILC2-derived IL-17 secreting cells isolated using an IL-17-secretion assay from blood ILC2s cultured with IL-1β, IL-23 and TGF-β for 5 days (see material and methods). IL-17 secreting ILCs were culture with IL-1β and IL-4 or in the presence of IL-1β, IL-23 and TGF-β for 5 days. Intracellular IL-5 and IL-17 was measured after restimulation with PMA and ionomycin. Data are representative of 2 experiments with independent donors (left). Number in quadrants and gates indicate percentage of cells in each. Bar graphs are the quantification of 2 independent experiments with one donor each.
Supplementary Figure 3 c-Kit and CCR6 identify a RORγt+ ILC2 subset which can readily transdifferentiate into IL-17 producing cells.
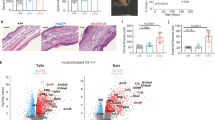
(a) Flow cytometry analysis of blood ILC2s with CCR6, RORγt and c-Kit, and gating strategy to isolate c-Kit+CCR6+ ILC2s, c-Kit− ILC2s, CD56bright NK cells and CRTH2−c-Kit+ ILCs, from human peripheral blood. Numbers adjacent to outlined areas indicate percent of each cell. Data are representative of 6 experiments with independent donors. (b) Heatmap display of expression levels of ILC subsets defining genes in c-Kit+CCR6+ ILC2s (n = 8), c-Kit− ILC2s (n = 8), CRTH2−c-Kit+ ILCs (n = 5) and CD56bright NK cells (n = 10) from blood and ILC1s (n = 5), NKp44+ ILC3s (n = 6) and NKp44− ILC3s (n = 6, all independent donors) from tonsils determined by RT-qPCR.. Subsets of ILCs were isolated as in Supplementary Fig. 3a. Red and blue doted box indicate ILC3- and ILC2- specifying genes, respectively. (c) Bar plots of expression level of selected genes from (b) in blood c-Kit+CCR6+ ILC2s, c-Kit− ILC2s, CRTH2−c-Kit+ ILCs and CD56bright NK cells. NS, not significant. 1-way ANOVA followed by Tukey’s test; * P < 0.05, ** P < 0.01, *** P < 0.001 and **** P< 0.0001. Error bars represent SEM. (d) Flow cytometry analyses of IL-13 production in naive, uncultured ILCs from blood stimulated with PMA and ionomycin (left) and dot plots for quantification of the frequency of IL-13 producing cells to parental cells (n = 3, independent donors). Numbers adjacent to outlined areas indicate percent IL-13+ cells. (e) Cloning strategy for differentiation of blood-derived ILC2s into ILC3s with IL-2, IL-23, TGF-β and IL-1β in the presence of feeder cells (irradiated allogeneic PBMCs and JY-cells) in 96-well plates. (f) Quantification of the number of c-Kit+CCR6+ ILC2s (n = 8), c-Kit− ILC2s (n = 7), CRTH2−c-Kit+ ILCs (n = 6) and CD56bright NK cells (n = 6, all independent donors) recovered from one well (among an initial 3 × 102 cells per well plated at day 0) after 7 d of culture with IL-1β, IL-25 or IL-33. 1-way ANOVA followed by Dunnet’s test (vs control condition, eg. no cytokines); ** P < 0.01. Error bars represent SEM. (g) Concentration of IL-5 in supernatants of c-Kit+CCR6+ ILC2s, c-Kit− ILC2s (key) plated as in b and cultured for 4 d with IL-1β, IL-25 or IL-33 (n = 4, independent donors). 1-way ANOVA followed by Dunnett’s test (vs control condition, eg. no cytokines); ** P < 0.01, *** P < 0.001 and **** P< 0.0001. Error bars represent SEM.
Supplementary Figure 4 TGF-β regulates the transition of c-Kit− ILC2s into RORγt+ ILC2s.
(a) Principle component analysis of global gene expression of c-Kit+ ILC2 and c-Kit− ILC2 populations isolated from 3 healthy donors, stimulated with indicated cytokines. c-Kit+ ILC2s (grey) and c-Kit− ILC2s (dotted circle) incubated under different cytokine combination (dot shown in different colors as indicated) are displayed in PC1 vs PC2 (top) and in PC1 vs PC3 (bottom) are shown. (b) Venn diagrams comparing differently expressed genes of c-Kit+ ILC2s and c-Kit− ILC2s stimulated with indicated cytokines from global gene expression data in (a). Red circle represents gene-expression differences of c-Kit+ ILC2s stimulated with IL-1β vs IL-1β IL-23. The blue circle represents differences of c-Kit− ILC2 stimulated with IL-1β vs IL-1β plus IL-23. Differently expressed genes of c-Kit− ILC2 stimulated with IL-1β vs IL-23 or IL-1β, IL-23 and TGF-β are shown in the green circle. Numbers in the Venn diagram represent numbers of differently expressed genes. (c) Expression of GATA3, IL17RB, IL1RL1 and CRLF2 determined by RT-qPCR in c-Kit+ ILC2s (n = 8) and c-Kit− ILC2s (n = 6, all independent donors) stimulated with IL-1β or IL-1β TGF-β for 7 days. ns, not significant. * P < 0.05, ** P < 0.01, *** P < 0.001 and **** P< 0.0001. 1-way ANOVA followed by Tukey’s test. Error bars represent SEM. (d) Flow cytometry analysis of RORγt and GATA-3 expression in c-Kit+ ILC2s, c-Kit− ILC2s, CD56bright NK and CRTH2−c-Kit+ ILCs stimulated with IL-1β or IL-1β TGF-β for 7 days. Data are representative of 3 independent experiments with individual donors. Numbers adjacent to outlined areas indicate percent of each cell.
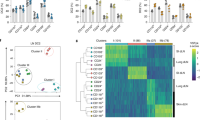
Supplementary Figure 5 Single cell RNAseq analysis identifies skin-homing RORγt+ ILC2s in peripheral blood.
(a) Representative differentially expressed genes among all cluster identified. Dot size represents the fraction of cells in the cluster expressing the gene and the color code shows average expression levels of the gene. (b) Overlay visualization of GATA3 and RORA expression on tSNE plots of single cell RNAseq data of ILC populations isolated from peripheral blood from one donor. (c, d) Expression of genes identified as differentially expressed genes between cluster 1 and cluster 2 in single cell RNAseq (c) and ILC2-related genes (d), determined by RT-qPCR in sort purified c-Kit+CCR10+ ILC2s, c-Kit+ ILC2s, c-Kit− ILC2s, CD56bright NK and CRTH2−c-Kit+ ILCs (all n = 4, independent donors). Data are shown in box-whisker plots. Line and “+” represent median and mean. Whiskers show min to max value. 1-way ANOVA followed by Dunnett’s test; * P < 0.05, ** P < 0.01, *** P < 0.001 and **** P< 0.0001; ns, not significant.
Supplementary information
Supplementary Information
Supplementary Figs. 1–5 and Supplementary Tables 1–4
Rights and permissions
About this article
Cite this article
Bernink, J.H., Ohne, Y., Teunissen, M.B.M. et al. c-Kit-positive ILC2s exhibit an ILC3-like signature that may contribute to IL-17-mediated pathologies. Nat Immunol 20, 992–1003 (2019). https://doi.org/10.1038/s41590-019-0423-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-019-0423-0
This article is cited by
-
Steady-state estradiol triggers a unique innate immune response to allergen resulting in increased airway resistance
Biology of Sex Differences (2023)
-
Innate lymphoid cells and innate-like T cells in cancer — at the crossroads of innate and adaptive immunity
Nature Reviews Cancer (2023)
-
Reciprocal costimulatory molecules control the activation of mucosal type 3 innate lymphoid cells during engagement with B cells
Cellular & Molecular Immunology (2023)
-
Osteoimmunology in Periodontitis and Orthodontic Tooth Movement
Current Osteoporosis Reports (2023)
-
Heterogeneity of type 2 innate lymphoid cells
Nature Reviews Immunology (2022)