Abstract
The helicase RIG-I initiates an antiviral immune response after recognition of pathogenic RNA. TRIM25, an E3 ubiquitin ligase, mediates K63-linked ubiquitination of RIG-I, which is crucial for RIG-I downstream signaling and the antiviral innate immune response. The components and mode of the RIG-I-initiated innate signaling remain to be fully understood. Here we identify a novel long noncoding RNA (Lnczc3h7a) that binds to TRIM25 and promotes RIG-I-mediated antiviral innate immune responses. Depletion of Lnczc3h7a impairs RIG-I signaling and the antiviral innate response to RNA viruses in vitro and in vivo. Mechanistically, Lnczc3h7a binds to both TRIM25 and activated RIG-I, serving as a molecular scaffold for stabilization of the RIG-I–TRIM25 complex at the early stage of viral infection. Lnczc3h7a facilitates TRIM25-mediated K63-linked ubiquitination of RIG-I and thus promotes downstream signaling transduction. Our findings reveal that host RNAs can enhance the response of innate immune sensors to foreign RNAs, ensuring effective antiviral defense.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon request. The RNA sequencing data from this study are deposited in NCBI GEO under accession code GSE112175.
Code availability
The code that supports the findings of this study is available from the corresponding author upon request.
References
Yoneyama, M. et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5, 730–737 (2004).
Hornung, V. et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314, 994–997 (2006).
Goubau, D. et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature 514, 372–375 (2014).
Chow, K. T., Gale, M. Jr. & Loo, Y.-M. RIG-I and other RNA sensors in antiviral immunity. Annu. Rev. Immunol. 36, 667–694 (2018).
Liu, J., Qian, C. & Cao, X. Post-translational modification control of innate immunity. Immunity 45, 15–30 (2016).
Chen, W. et al. Induction of Siglec-G by RNA viruses inhibits the innate immune response by promoting RIG-I degradation. Cell 152, 467–478 (2013).
Zhao, K. et al. Cytoplasmic STAT4 promotes antiviral type I IFN production by blocking CHIP-mediated degradation of RIG-I. J. Immunol. 196, 1209–1217 (2016).
Gack, M. U. et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446, 916–920 (2007).
Gack, M. U. et al. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc. Natl Acad. Sci. USA 105, 16743–16748 (2008).
Jiang, X. et al. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity 36, 959–973 (2012).
Manokaran, G. et al. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 350, 217–221 (2015).
Choudhury, N. R. et al. RNA-binding activity of TRIM25 is mediated by its PRY/SPRY domain and is required for ubiquitination. BMC Biol. 15, 105 (2017).
Sanchez, J. G. et al. TRIM25 binds RNA to modulate cellular anti-viral defense. J. Mol. Biol. 430, 5280–5293 (2018).
Chen, Y. G., Satpathy, A. T. & Chang, H. Y. Gene regulation in the immune system by long noncoding RNAs. Nat. Immunol. 18, 962–972 (2017).
Zhang, Y. & Cao, X. Long noncoding RNAs in innate immunity. Cell. Mol. Immunol. 13, 138–147 (2015).
Liu, B. et al. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat. Immunol. 18, 499–508 (2017).
Wang, P. et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 344, 310–313 (2014).
Wang, P., Xu, J., Wang, Y. & Cao, X. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science 358, 1051–1055 (2017).
Jiang, M. et al. Self-recognition of an inducible host lncRNA by RIG-I feedback restricts innate immune response. Cell 173, 906–919.e913 (2018).
Lin, M. F., Jungreis, I. & Kellis, M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27, i275–282 (2011).
Sanchez, J. G. et al. The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer. Proc. Natl Acad. Sci. USA 111, 2494–2499 (2014).
Koliopoulos, M. G., Esposito, D., Christodoulou, E., Taylor, I. A. & Rittinger, K. Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity. EMBO J. 35, 1204–1218 (2016).
Pauli, E.-K. et al. The ubiquitin-specific protease USP15 promotes RIG-I-mediated antiviral signaling by deubiquitylating TRIM25. Sci. Signal. 7, ra3–ra3 (2014).
Inn, K.-S. et al. Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Mol. Cell 41, 354–365 (2011).
Castanier, C. et al. MAVS ubiquitination by the E3 ligase TRIM25 and degradation by the proteasome is involved in type I interferon production after activation of the antiviral RIG-I-like receptors. BMC Biol. 10, 44 (2012).
McNab, F., Mayer-Barber, K., Sher, A., Wack, A. & O’Garra, A. Type I interferons in infectious disease. Nat. Rev. Immunol. 15, 87–103 (2015).
Liu, S. et al. Nuclear RNF2 inhibits interferon function by promoting K33-linked STAT1 disassociation from DNA. Nat. Immunol. 19, 41–52 (2018).
Peisley, A., Wu, B., Xu, H., Chen, Z. J. & Hur, S. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature 509, 110–114 (2014).
Seth, R. B., Sun, L., Ea, C.-K. & Chen, Z. J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell 122, 669–682 (2005).
Lu, C. et al. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure 18, 1032–1043 (2010).
Kowalinski, E. et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 147, 423–435 (2011).
Cui, S. et al. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell 29, 169–179 (2008).
Lässig, C. et al. ATP hydrolysis by the viral RNA sensor RIG-I prevents unintentional recognition of self-RNA. eLife 4, e10859 (2015).
Louber, J., Brunel, J., Uchikawa, E., Cusack, S. & Gerlier, D. Kinetic discrimination of self/non-self RNA by the ATPase activity of RIG-I and MDA5. BMC Biol. 13, 54 (2015).
Wang, K. C. & Chang, H. Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914 (2011).
Atianand, M. K. et al. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell 165, 1672–1685 (2016).
Guttman, M. et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300 (2011).
Collins, K. Physiological assembly and activity of human telomerase complexes. Mech. Ageing Dev. 129, 91–98 (2008).
Zhao, J., Sun, B. K., Erwin, J. A., Song, J.-J. & Lee, J. T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322, 750–756 (2008).
Yoon, J.-H. et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat. Commun. 4, 2939 (2013).
Fu, M. & Blackshear, P. J. RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat. Rev. Immunol. 17, 130–143 (2016).
Treiber, T. et al. A compendium of RNA-binding proteins that regulate microRNA biogenesis. Mol. Cell 66, 270–284.e213 (2017).
Liang, J., Song, W., Tromp, G., Kolattukudy, P. E. & Fu, M. Genome-wide survey and expression profiling of CCCH-zinc finger family reveals a functional module in macrophage activation. PLoS ONE 3, e2880 (2008).
Chiang, J. J. et al. Viral unmasking of cellular 5S rRNA pseudogene transcripts induces RIG-I-mediated immunity. Nat. Immunol. 19, 53–62 (2018).
Wang, W. et al. RNF122 suppresses antiviral type I interferon production by targeting RIG-I CARDs to mediate RIG-I degradation. Proc. Natl Acad. Sci. USA 113, 9581–9586 (2016).
Huppertz, I. et al. iCLIP: protein–RNA interactions at nucleotide resolution. Methods 65, 274–287 (2014).
Acknowledgements
This work is supported by grants from the National Natural Science Foundation of China (81788101, 31390431 and 81871236), National 135 Mega Program of China (2017ZX10102032-001, 2017ZX10202203-002), National Key Research and Development Program of China (2018YFA0507403) and CAMS Innovation Fund for Medical Sciences (2016-12M-1-003).
Author information
Authors and Affiliations
Contributions
H.L., M.J., L.L., Z.Y., Z.M. and S.L. performed the experiments. Y.M. and L.Z. generated knockout RAW264.7 cell lines and mice. X.C., H.L. and M.J. analyzed data and wrote the manuscript. X.C. was responsible for research supervision, coordination and strategy.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
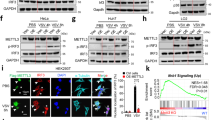
Supplementary Fig. 1 Characterization of Lnczc3h7a.
a, RNAi screen of the top 20 abundance lncRNAs co-precipitated with Flag-TRIM25: qRT-PCR analysis of Ifnb1 expression in peritoneal macrophages transfected with different siRNAs targeting these lncRNAs for 48 hr, then infected with VSV for 8 hr. b, 5’ and 3’ RACE assay of Lnczc3h7a in peritoneal macrophages upon VSV infection for 8 hr, using the primer of 5’RACE and 3’RACE (top). Schematic illustration of Lnczc3h7a locus on the introns of Zc3h7a and primers (labeled with arrows) used in RACE assay (below). c, Schematic illustration of different nucleotides between Lnczc3h7a and Zc3h7a intron. d, Schematic representation of PacBio long-read sequencing data shown that no transcript locus on the homologous region of Lnczc3h7a in A549 cell genome after VSV infection for 8 hr. e, Schematic illustration of primers (labeled with arrows) used in RACE assay which targeted the homologous region of 5’ end 291 bp sequence in human genome (top) and 5’ and 3’ RACE assay of equivalent Lnczc3h7a in A549 cells upon VSV infection for 8 hr (below). Data are representative of three independent experiments with n = 3 technical replicates (a), three independent experiments (b-e), each symbol represents an individual technical replicate (a) (shown as mean and s.e.m. in a), two-tailed unpaired Student’s t-test.
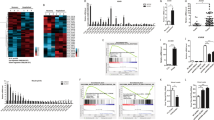
Supplementary Fig. 2 Lnczc3h7a is a non-coding RNA.
a, Ribosome sedimentation analysis of Gapdh, 18S, 28S and Lnczc3h7a from peritoneal macrophages infected with VSV for 8 hr. Cell lysates were fractionated by sucrose gradient centrifugation. b, Prediction of putative proteins encoded by Lnczc3h7a (red rectangle) using ORF Finder (http://www.ncbi.nlm.nih.gov/orffinder/). c, The CSF scores of Lnczc3h7a determined by analysis with PhyloCSF. Data are representative of three independent experiments (a).
Supplementary Fig. 3 Lnczc3h7a levels are upregulated by innate immune stimulators and are high in immune-related organs and immune cells.
a, qRT-PCR analysis of Lnczc3h7a and Isg54 expression in peritoneal macrophages stimulated with several types of stimulators for indicated hours. b, Northern blot analysis of Lnczc3h7a from peritoneal macrophages infected with VSV for 8 hr. c, qRT-PCR analysis of Lnczc3h7a expression in indicated organs. d, qRT-PCR analysis of Lnczc3h7a expression in indicated cells. Peritoneal macrophages were isolated from the peritoneal cavities of mice 3 days after injection with thioglycolate medium. BMDCs were isolated and induced from wild type mice’s bone marrow. CD4+ T cells, CD8+ T cells, NK cells and B cells were isolated from wild type mice’s spleens. Data are representative of three independent experiments with n = 3 technical replicates (a,c,d), three independent experiments (b), each symbol represents an individual technical replicate (a,c,d) (shown as mean and s.e.m. in a,c,d), two-tailed unpaired Student’s t-test.
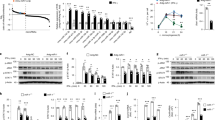
Supplementary Fig. 4 Knockdown or knockout of Lnczc3h7a blocks RIG-I-triggered antiviral signaling pathway.
a, Schematic illustration of the locations of si-Lnczc3h7a target sequence. b, qRT-PCR analysis of Lnczc3h7a and Zc3h7a expression in peritoneal macrophages transfected with si-Ctrl or si-Lnczc3h7a for 48 hr and then infected with VSV for 8 hr. c, Schematic illustration of the knockout region of Lnczc3h7a-/- RAW264.7 cells with two sgRNAs and test primers labeled with arrows. d, PCR analysis of genome deletion in Lnczc3h7a-/- and Lnczc3h7a+/+ RAW264.7 cells. e, qRT-PCR analysis of Zc3h7a expression in Lnczc3h7a-/- and Lnczc3h7a+/+ RAW264.7 cells infected with VSV for 8 hr. f, Immunoblot analysis of RIG-I signaling pathway in peritoneal macrophages transfected with si- Ctrl or si-Lnczc3h7a for 48 hr and infected with VSV for indicated hours. Data are representative of three independent experiments with n = 3 technical replicates (b,e), three independent experiments (d,f), each symbol represents an individual technical replicate (b,e) (shown as mean and s.e.m. in b,e), two-tailed unpaired Student’s t-test.
Supplementary Fig. 5 Lnczc3h7a promotes antiviral innate responses in mice.
a, Schematic illustration of the knockout region of Lnczc3h7a-/- mice with two sgRNAs and test primers labeled with arrows. b, PCR analysis of genome deletion in Lnczc3h7a+/+, Lnczc3h7a+/- and Lnczc3h7a-/- mice. c, qRT-PCR analysis of Zc3h7a expression in peritoneal macrophages from Lnczc3h7a+/+and Lnczc3h7a-/- mice, infected with VSV for 8 hr. d, ELISA of IFN-α, IFN-β and IL-6 in the supernatants of peritoneal macrophages from Lnczc3h7a+/+ and Lnczc3h7a-/- mice, infected with VSV for 12 hr. e, Immunoblot analysis of RIG-I signaling pathway in peritoneal macrophages from Lnczc3h7a+/+ and Lnczc3h7a-/- mice, infected with VSV for indicated hours. f, ELISA of the production of IFN-α, IFN-β and IL-6 in sera from Lnczc3h7a+/+and Lnczc3h7a-/- mice at 24 hr after intranasal injection with influenza A virus (100 pfu per mouse). g, ELISA of the production of IFN-α, IFN-β and IL-6 in sera from Lnczc3h7a+/+and Lnczc3h7a-/- mice at 18 hr after i.v. injection with HSV-1 virus (1×107 pfu/g). h, qRT-PCR analysis of Ifna, Ifnb1, Il6 and Ifit1 mRNA levels in organs from g. i, Hematoxylin-and eosin staining of lung sections from mice in g. Scale bar, 50 μm. Data are representative of three independent experiments (b,e,i), three independent experiments with n = 3 technical replicates (c,d,f-h), each symbol represents an individual technical replicate (c,d,f-h) (shown as mean and s.e.m. in c,d,f-h), two-tailed unpaired Student’s t-test.
Supplementary Fig. 6 Lnczc3h7a facilitates RIG-I signaling pathway by promoting the interaction of TRIM25 and RIG-I.
a, Co-immunoprecipitation analysis of oligomerization and K48-linked ubiquitination of TRIM25 in HEK293T cells co-transfected with flag-TRIM25, V5-TRIM25, HA-Ub-K48 and Lnczc3h7a for 24 hr, infected with VSV for 8 hr. IP, immunoprecipitation; WCL, whole cell lysates. b, Co-immunoprecipitation analysis of the interaction of TRIM25 and MAVS and K48-linked ubiquitination of MAVS in HEK 293T cells co-transfected with flag-TRIM25, V5-MAVS, HA-Ub-K48 and Lnczc3h7a for 24 hr, infected with VSV for 8 hr. c, Co-immunoprecipitation analysis of the interaction of TRIM25 with RIG-I and K63-linked ubiquitination of RIG-I in HEK 293T cells co-transfected with flag-RIG-I, V5-TRIM25, HA-Ub-K63 and Lnczc3h7a for 24 hr, infected with VSV for 8 hr. d, Immunofluorescence assay of the co-localization of endogenous K63-Ub (green) and RIG-I (red) in Lnczc3h7a+/+ and Lnczc3h7a-/- mice peritoneal macrophages upon VSV infection for 8 hr. Nuclei were stained with DAPI (blue). Scale bar, 5 μm. e, Co-immunoprecipitation analysis of the interaction of RIG-I and MAVS in the presence of TRIM25 or Lnczc3h7a in HEK 293T cells co-transfected with flag-RIG-I, V5-MAVS, Myc-TRIM25 and Lnczc3h7a for 24 hr, and infected with VSV for 8 hr. f. Schematic illustration of the location of Trim25 sgRNA target sequence (top) and frameshift mutation sequence (below). Data are representative of three independent experiments (a-e).
Supplementary Fig. 7 Lnczc3h7a binds to TRIM25 and RIG-I at nucleotide 308, 311 and 332 during viral infection.
a, Predicted RNA secondary structure of Lnczc3h7a based on minimum free energy (MFE) (http://rna.tbi.univie.ac.at/). b, Predicted RNA secondary structure of three control transcripts based on MFE (http://rna.tbi.univie.ac.at/). c, Co-immunoprecipitation analysis of the interaction of TRIM25 with RIG-I and K63-linked ubiquitination of RIG-I in Lnczc3h7a-/- L929 cells co-transfected with Flag-TRIM25, V5-RIG-I, HA-Ub-K63 and three control RNAs or Lnczc3h7a for 24 hr, infected with VSV for 8 hr. d, Co-immunoprecipitation analysis of the interaction of TRIM25 with RIG-I and K63-linked ubiquitination of RIG-I in Lnczc3h7a-/- L929 cells co-transfected with Flag-TRIM25, V5-RIG-I, HA-Ub-K63 and different Lnczc3h7a delete mutants for 24 hr, infected with VSV for 8 hr. e, Immunofluorescence analysis of the co-localization of TRIM25 (red) and RIG-I (green)in Lnczc3h7a-/- L929 cells co-transfected with Flag-TRIM25, V5-RIG-I, HA-Ub-K63 and different Lnczc3h7a delete mutants for 24 hr, infected with VSV for 8 hr. Nuclei were stained with DAPI (blue). Scale bar, 5 μm. Data are representative of three independent experiments (c-e).
Supplementary information
Supplementary Tables 1-6
the top 20 abundance lncRNAs with RIP–sequencing
Rights and permissions
About this article
Cite this article
Lin, H., Jiang, M., Liu, L. et al. The long noncoding RNA Lnczc3h7a promotes a TRIM25-mediated RIG-I antiviral innate immune response. Nat Immunol 20, 812–823 (2019). https://doi.org/10.1038/s41590-019-0379-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-019-0379-0
This article is cited by
-
TRIM25 promotes glioblastoma cell growth and invasion via regulation of the PRMT1/c-MYC pathway by targeting the splicing factor NONO
Journal of Experimental & Clinical Cancer Research (2024)
-
Chromosome-level genome provides insights into environmental adaptability and innate immunity in the common dolphin (delphinus delphis)
BMC Genomics (2024)
-
Disparate macrophage responses are linked to infection outcome of Hantan virus in humans or rodents
Nature Communications (2024)
-
LncRNA INHEG promotes glioma stem cell maintenance and tumorigenicity through regulating rRNA 2’-O-methylation
Nature Communications (2023)
-
Differential expression profile and in-silico functional analysis of long noncoding RNA and mRNA in duck embryo fibroblasts infected with duck plague virus
BMC Genomics (2022)