Abstract
Human inborn errors of IFN-γ immunity underlie mycobacterial diseases. We describe patients with Mycobacterium bovis (BCG) disease who are homozygous for loss-of-function mutations of SPPL2A. This gene encodes a transmembrane protease that degrades the N-terminal fragment (NTF) of CD74 (HLA invariant chain) in antigen-presenting cells. The CD74 NTF therefore accumulates in the HLA class II+ myeloid and lymphoid cells of SPPL2a-deficient patients. This toxic fragment selectively depletes IL-12- and IL-23-producing CD1c+ conventional dendritic cells (cDC2s) and their circulating progenitors. Moreover, SPPL2a-deficient memory TH1* cells selectively fail to produce IFN-γ when stimulated with mycobacterial antigens in vitro. Finally, Sppl2a–/– mice lack cDC2s, have CD4+ T cells that produce small amounts of IFN-γ after BCG infection, and are highly susceptible to infection with BCG or Mycobacterium tuberculosis. These findings suggest that inherited SPPL2a deficiency in humans underlies mycobacterial disease by decreasing the numbers of cDC2s and impairing IFN-γ production by mycobacterium-specific memory TH1* cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Bustamante, J. et al. Mendelian susceptibility to mycobacterial disease: genetic, immunological, and clinical features of inborn errors of IFN-gamma immunity. Semin. Immunol. 26, 454–470 (2014).
Casanova, J. L. & Abel, L. Genetic dissection of immunity to mycobacteria: the human model. Annu. Rev. Immunol. 20, 581–620 (2002).
de Beaucoudrey, L. et al. Revisiting human IL-12Rbeta1 deficiency: a survey of 141 patients from 30 countries. Medicine 89, 381–402 (2010).
Dupuis, S. et al. Human interferon-gamma-mediated immunity is a genetically controlled continuous trait that determines the outcome of mycobacterial invasion. Immunol. Rev. 178, 129–137 (2000).
Kreins, A. Y. et al. Human TYK2 deficiency: mycobacterial and viral infections without hyper-IgE syndrome. J. Exp. Med. 212, 1641–1662 (2015).
Bogunovic, D. et al. Mycobacterial disease and impaired IFN-gamma immunity in humans with inherited ISG15 deficiency. Science 337, 1684–1688 (2012).
Bustamante, J. et al. Germline CYBB mutations that selectively affect macrophages in kindreds with X-linked predisposition to tuberculous mycobacterial disease. Nat. Immunol. 12, 213–221 (2011).
Boisson-Dupuis, S. et al. Inherited and acquired immunodeficiencies underlying tuberculosis in childhood. Immunol. Rev. 264, 103–120 (2015).
Nathan, C. F. et al. Identification of interferon-gamma as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J. Exp. Med. 158, 670–689 (1983).
Prando, C. et al. Inherited IL-12p40 deficiency: genetic, immunologic, and clinical features of 49 patients from 30 kindreds. Medicine 92, 109–122 (2013).
Kong, X. F. et al. A novel homozygous p.R1105X mutation of the AP4E1 gene in twins with hereditary spastic paraplegia and mycobacterial disease. PLoS One 8, e58286 (2013).
Itan, Y. et al. The mutation significance cutoff: gene-level thresholds for variant predictions. Nat. Methods 13, 109–110 (2016).
Voss, M. et al. Mechanism, specificity, and physiology of signal peptide peptidase (SPP) and SPP-like proteases. Biochim. Biophys. Acta 1828, 2828–2839 (2013).
Brady, O. A. et al. Regulated intramembrane proteolysis of the frontotemporal lobar degeneration risk factor, TMEM106B, by signal peptide peptidase-like 2a (SPPL2a). J. Biol. Chem. 289, 19670–19680 (2014).
Fluhrer, R. et al. A gamma-secretase-like intramembrane cleavage of TNFα by the GxGD aspartyl protease SPPL2b. Nat. Cell Biol. 8, 894–896 (2006).
Friedmann, E. et al. SPPL2a and SPPL2b promote intramembrane proteolysis of TNFα in activated dendritic cells to trigger IL-12 production. Nat. Cell Biol. 8, 843–848 (2006).
Kirkin, V. et al. The Fas ligand intracellular domain is released by ADAM10 and SPPL2a cleavage in T-cells. Cell Death Differ. 14, 1678–1687 (2007).
Martin, L. et al. Regulated intramembrane proteolysis of Bri2 (Itm2b) by ADAM10 and SPPL2a/SPPL2b. J. Biol. Chem. 283, 1644–1652 (2008).
Beisner, D. R. et al. The intramembrane protease Sppl2a is required for B cell and DC development and survival via cleavage of the invariant chain. J. Exp. Med. 210, 23–30 (2013).
Bergmann, H. et al. B cell survival, surface BCR and BAFFR expression, CD74 metabolism, and CD8- dendritic cells require the intramembrane endopeptidase SPPL2A. J. Exp. Med. 210, 31–40 (2013).
Schneppenheim, J. et al. The intramembrane protease SPPL2a promotes B cell development and controls endosomal traffic by cleavage of the invariant chain. J. Exp. Med. 210, 41–58 (2013).
Fleck, D. et al. Proteolytic processing of neuregulin 1 type III by three intramembrane-cleaving proteases. J. Biol. Chem. 291, 318–333 (2016).
Itan, Y. et al. The human gene connectome as a map of short cuts for morbid allele discovery. Proc. Natl. Acad. Sci. USA 110, 5558–5563 (2013).
Zhang, Y. et al. Genomics is rapidly advancing precision medicine for immunological disorders. Nat. Immunol. 16, 1001–1004 (2015).
Rieux-Laucat, F. Inherited and acquired death receptor defects in human autoimmune lymphoproliferative syndrome. Curr. Dir. Autoimmun. 9, 18–36 (2006).
Ai, J. W. et al. The risk of tuberculosis in patients with rheumatoid arthritis treated with tumor necrosis factor-alpha antagonist: a metaanalysis of both randomized controlled trials and registry/cohort studies. J. Rheumatol. 42, 2229–2237 (2015).
Moreno-De-Luca, A. et al. Adaptor protein complex-4 (AP-4) deficiency causes a novel autosomal recessive cerebral palsy syndrome with microcephaly and intellectual disability. J. Med. Genet. 48, 141–144 (2011).
Schneppenheim, J. et al. Signal-peptide-peptidase-like 2a is required for CD74 intramembrane proteolysis in human B cells. Biochem. Biophys. Res. Commun. 451, 48–53 (2014).
Becker-Herman, S. et al. CD74 is a member of the regulated intramembrane proteolysis-processed protein family. Mol. Biol. Cell 16, 5061–5069 (2005).
Hambleton, S. et al. IRF8 mutations and human dendritic-cell immunodeficiency. N. Engl. J .Med. 365, 127–138 (2011).
Bridgeford, E. C. et al. Agammaglobulinemia and Staphylococcus aureus botryomycosis in a cohort of related sentinel Swiss Webster mice. J. Clin. Microbiol. 46, 1881–1884 (2008).
Conley, M. E. et al. Primary B cell immunodeficiencies: comparisons and contrasts. Annu. Rev. Immunol. 27, 199–227 (2009).
Reynolds, G. & Haniffa, M. Human and mouse mononuclear phagocyte networks: a tale of two species? Front. Immunol. 6, 330 (2015).
Merad, M. et al. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 31, 563–604 (2013).
Iwakoshi, N. N. et al. The transcription factor XBP-1 is essential for the development and survival of dendritic cells. J. Exp. Med. 204, 2267–2275 (2007).
Dickinson, R. E. et al. The evolution of cellular deficiency in GATA2 mutation. Blood 123, 863–874 (2014).
Bigley, V. et al. Dendritic cell analysis in primary immunodeficiency. Curr. Opin. Allergy. Clin. Immunol. 16, 530–540 (2016).
Vignali, D. A. & Kuchroo, V. K. IL-12 family cytokines: immunological playmakers. Nat. Immunol. 13, 722–728 (2012).
See, P. et al. Mapping the human DC lineage through the integration of high-dimensional techniques. Science 356, eaag3009 (2017).
Villani, A. C. et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573 (2017).
Breton, G. et al. Circulating precursors of human CD1c+ and CD141+ dendritic cells. J. Exp. Med. 212, 401–413 (2015).
Ma, C. S. et al. Unique and shared signaling pathways cooperate to regulate the differentiation of human CD4+ T cells into distinct effector subsets. J. Exp. Med. 213, 1589–1608 (2016).
Teng, M. W. et al. IL-12 and IL-23 cytokines: from discovery to targeted therapies for immune-mediated inflammatory diseases. Nat. Med. 21, 719–729 (2015).
Acosta-Rodriguez, E. V. et al. Surface phenotype and antigenic specificity of human interleukin 17–producing T helper memory cells. Nat. Immunol. 8, 639–646 (2007).
Geiger, R. et al. Human naive and memory CD4+ T cell repertoires specific for naturally processed antigens analyzed using libraries of amplified T cells. J. Exp. Med. 206, 1525–1534 (2009).
Marquis, J. F. et al. Interferon regulatory factor 8 regulates pathways for antigen presentation in myeloid cells and during tuberculosis. PLoS Genet. 7, e1002097 (2011).
Berghout, J. et al. Irf8-regulated genomic responses drive pathological inflammation during cerebral malaria. PLoS Pathog. 9, e1003491 (2013).
Bigley, V. et al. The human syndrome of dendritic cell, monocyte, B and NK lymphoid deficiency. J. Exp. Med. 208, 227–234 (2011).
Kindler, V. et al. The inducing role of tumor necrosis factor in the development of bactericidal granulomas during BCG infection. Cell 56, 731–740 (1989).
Keane, J. et al. Tuberculosis associated with infliximab, a tumor necrosis factor alpha-neutralizing agent. N. Engl. J. Med. 345, 1098–1104 (2001).
Thiant, S. et al. Imatinib mesylate inhibits STAT5 phosphorylation in response to IL-7 and promotes T cell lymphopenia in chronic myelogenous leukemia patients. Blood Cancer J. 7, e551 (2017).
Martínez-Barricarte, R. et al. Transduction of Herpesvirus saimiri-transformed T cells with exogenous genes of interest. Curr. Protoc. Immunol. 115, 7.21C.1–7.21C.12 (2016).
Schneppenheim, J. et al. The intramembrane proteases signal peptide peptidase-like 2a and 2b have distinct functions in vivo. Mol. Cell Biol. 34, 1398–1411 (2014).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Wang, K. et al. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Med. Genet. 81, 559–575 (2007).
Abecasis, G. R. et al. Merlin: rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 30, 97–101 (2002).
Breton, G. et al. Defining human dendritic cell progenitors by multiparametric flow cytometry. Nat. Protoc. 10, 1407–1422 (2015).
Avery, D. T. et al. B cell-intrinsic signaling through IL-21 receptor and STAT3 is required for establishing long-lived antibody responses in humans. J. Exp. Med. 207, 155–171 (2010).
Lindestam Arlehamn, C. S. et al. A quantitative analysis of complexity of human pathogen-specific CD4 T cell responses in healthy M. tuberculosis infected South Africans. PLoS Pathog. 12, e1005760 (2016).
Gros, P. et al. Genetic control of natural resistance to Mycobacterium bovis (BCG) in mice. J. Immunol. 127, 2417–2421 (1981).
Langlais, D. et al. The macrophage IRF8/IRF1 regulome is required for protection against infections and is associated with chronic inflammation. J. Exp. Med. 213, 585–603 (2016).
Ng, S. L. et al. IκB kinase epsilon (IKK(epsilon)) regulates the balance between type I and type II interferon responses. Proc. Natl. Acad. Sci. USA 108, 21170–21175 (2011).
Mancino, A. et al. A dual cis-regulatory code links IRF8 to constitutive and inducible gene expression in macrophages. Genes Dev. 29, 394–408 (2015).
Barish, G. D. et al. Bcl-6 and NF-kappaB cistromes mediate opposing regulation of the innate immune response. Genes Dev. 24, 2760–2765 (2010).
Saliba, D. G. et al. IRF5:RelA interaction targets inflammatory genes in macrophages. Cell Rep. 8, 1308–1317 (2014).
Kaikkonen, M. U. et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol. Cell 51, 310–325 (2013).
Ostuni, R. et al. Latent enhancers activated by stimulation in differentiated cells. Cell 152, 157–171 (2013).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Thorvaldsdottir, H. et al. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013).
Acknowledgements
We thank B. Coller, C. Rice, X. Ma, M. Ciancanelli, and G. Vogt for helpful discussions and critical reading. We thank Y. Nemirovskaya, E. Anderson, T. Kochetkov, M. Romanick, L. Amar, C. Patissier, C. Desvallées, M. Woollett, D. Papandrea, A. Gall, and J. Gonzalez for technical and secretarial assistance, and all members of the Laboratory of Human Genetics of Infectious Diseases for helpful discussions. We thank F. Batteux and M. Bahuad from the Laboratory of Immunology, Cochin Hospital, Paris, France, for serological testing of patients, and F. B. Menozzi and the rest of the Microbiology Institute, EOC, Bellinzona, for providing microbial products. X.-F.K. was supported by the Jerome Lejeune Foundation, the Stony Wold-Herbert Fund, the Choh-Hao Li Memorial Fund Scholar Award, and the Shanghai Educational Development Foundation. R.M.B. was funded by a European Molecular Biology Organization (EMBO) long-term fellowship. J.M. was supported by the Charles H. Revson Senior Fellowship in Biomedical Sciences. B.S. received support from the Deutsche Forschungsgemeinschaft as part of the SFB877 and the Cluster of Excellence ‘Inflammation at Interfaces’, and the SCHR 1284/1-1 grant. The Laboratory of Human Genetics of Infectious Diseases is supported by grants from the St. Giles Foundation (J.-L.C.), The Rockefeller University Center for Clinical and Translational Science (grant UL1TR001866 from the National Center for Research Resources and the National Center for Advancing Sciences (NCATS) to R.M.-B. and X.-F.K.), the National Institutes of Health, the National Institute of Allergy and Infectious Diseases (5R01AI089970-02 and 5R37AI095983 to J.-L.C.), the Integrative Biology of Emerging Infectious Diseases Laboratory of Excellence (ANR-10-LABX-62-IBEID, and the French National Research Agency (ANR) under the ‘Investments for the future’ program (ANR-10-IAHU-01 to L.A.), ANR-IFNGPHOX (ANR-13-ISV3-0001-01, to J.B.), and ANR-GENMSMD (ANR-16-CE17-0005-01, to J.B.), Institut National de la Santé et de la Recherche Médicale (INSERM), Paris Descartes University, and The Rockefeller University. E.K.D., C.S.M., and S.G.T. are supported by research grants and fellowships from the National Health and Medical Research Council of Australia. D.L. was supported by a fellowship from the Fonds de Recherche du Québec Santé. Work in P.G.’s laboratory was supported by a grant from the National Institute of Allergy and Infectious Diseases (R01AI035237-19). The work at the Institute for Research in Biomedicine was supported by grants from the ERC (323183 PREDICT, to F.S.), the Swiss National Science Foundation (170213, to F.S.), and the Helmut Horten Foundation. This work was supported by award U19AI118626, NIH NIAID (to A.S. and F.S.).
Author information
Authors and Affiliations
Contributions
X.-F.K., R.M.-B., J.K., F.M., T. Lazarov, E.K.D., D. Langlais, C.S.M., G.B., T. Lasseau, L.A., C.T., S.B.-D., M.C.N., D. Latorre, J.M., B.S., K.L., G.R., S.G.T., F.G., F.S., P.G., J.B., and J.-L.C. designed experiments. X.-F.K., R.M.-B., F.M., J.K., T. Lasseau, E.K.D., D. Langlais, G.R., C.T., C.S.M., J.M., G.B., J.M., C.T., T. Lazarov, C.D., K.B.L., and D. Latorre conducted experiments. F.J.-H., Y.I., and L.A. performed bioinformatics analysis. X.-F.K., R.M.-B., F.M., J.K., T. Lazarov, E.K.D., D. Langlais, C.S.M., G.B., S.H., L.A., S.B.D., M.C.N., B.S., G.R., S.G.T., F.G., F.S., K.L., P.G., J.B., and J.-L.C. analyzed and/or interpreted data. A.B., C.A., C.P., K.A., D.M.-V., F.A., A.I., F.D., and I.B. provided samples and performed clinical diagnoses and follow-up of the patients. C.S.L.A., A.S., and B.S. contributed critical materials. J.B. and J.-L.C. supervised the study. X.-F.K., R.M.-B., and J.-L.C. wrote the manuscript and designed the figures, to which all other authors contributed. All authors edited the manuscript and approved its final version. X.-F.K. and R.M.-B. contributed equally. J.K., F.M., T. Lazarov, E.K.D., and C.S.M. contributed equally. G.B., K.B.L., and D. Langlais contributed equally. A.B. and C.A. contributed equally. B.S., M.C.N., and K.L. contributed equally. F.G., S.G.T., P.G., and F.S. contributed equally. J.B. and J.-L.C. contributed equally.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
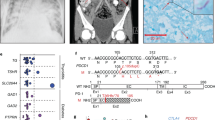
Supplementary Figure 1 Whole-genome linkage, whole-exome sequencing, population genetics, qPCR and effect of SPPL2a deficiency on TNF processing.
a) Combined whole-genome linkage analysis of kindreds A and B. LOD scores are shown in black and blue for alternating chromosomes, and information content is reported as a red trace. SPPL2A maps to the linked region of chromosome 15 indicated with a red arrow. b) The upper panel shows the IGV (https://www.broadinstitute.org/igv/) viewer presentation of the region of the genome containing the mutation for the index cases of both kindreds, in an antisense direction. Mutations are located in the center and the alternative nucleotide is indicated in red. The lower panels show the Sanger sequencing results for all the patients in each family, heterozygous carriers and one WT individual for each of the variants. The mutation site is marked with a dashed square. The shaded squares on the electrophoretograms represent the exonic areas, the rest corresponding to introns. c) Graph of CADD score on the y axis against minor allele frequency (MAF) on the x-axis, for all the heterozygous SPPL2A variants reported in the gnomAD database (http://gnomad.broadinstitute.org). The horizontal dashed line represents the MSC for SPPL2A. d, e) qPCR on cDNA from: d) EBV-B cells from a healthy control, a heterozygous carrier and P1 e) SV40 fibroblasts from a healthy control, a heterozygous carrier and P2 using a probe spanning exons 2-3 of SPPL2A. Mean and SEM is represented in both graphs. f) Schematic representation of the structure and cleavage sites of TNF. The SPPL2a and SPPL2b cleavage site in the transmembrane domain (indicated in black) is mark as a red dashed line. g) Memory and h) naive CD4+ T cells were isolated and stimulated with T cell activation and expansion (TAE) beads alone or under TH1 (TAE beads+IL-12) polarizing conditions. TNF secretion was assessed after and then the cells were re-stimulated with PMA/ionomycin and the percentage of TNF-positive cells was determined, by intracellular staining and flow cytometry. Mean is shown in both graphs.
Supplementary Figure 2 Intracellular versus surface expression of CD74 in B cells, T cells, and monocytes, and flow cytometry gating strategy.
a) Gating strategy used for (b) and (c). b) Representative histogram plots from Fig. 2f showing intracellular CD74 staining for B cells, monocytes and T cells from a healthy control (black), a heterozygous individual (blue) and a SPPL2a-deficient patient (red). c) Representative histogram plots from Fig. 2g showing surface CD74 staining for B cells, monocytes and T cells from a healthy control (black), a heterozygous individual (blue) and a SPPL2a-deficient patient (red). d) Gating strategy for Fig. 2h and 5a. PBMCs were depleted of the populations indicated in the curved arrows by MACS. These populations were analyzed with the gating strategy shown in the panels. After depletion of the CD14+, CD3+, CD19+ and CD56+ populations, the flow-through was analyzed with the markers shown in the gating strategy for analysis of the different DC populations.
Supplementary Figure 3 PBMCs from four healthy controls (WT/WT), two heterozygous carriers (WT/M) and three patients (M/M) were studied.
a) Gating strategy to identify B cell subsets. b) Representative data from FACS analysis for B cell subsets obtained with CD19, CD20, CD10 and CD27 Abs, in WT and SPPL2a-deficient patients. c) Percentages of memory B cells from healthy controls (WT/WT), a healthy WT/WT family member, heterozygous carriers (WT/M) and patients with AR SPPL2a deficiency (M/M) expressing IgG (left panel) or IgA (right panel). Means are indicated with a black line. d). Whiskers (5-95 percentile) graph of the ex vivo IgM, IgG and IgA production by naive and memory B cells from healthy controls, patients and heterozygous carriers following stimulation with CD40L and IL-21. e) BAFF-R expression (upper panel) on naive and memory B cells and HLA-DR expression on monocytes and B cells (lower panel) were assessed by FACS and are presented relative to the results for healthy controls; there were no significant differences between healthy donors, heterozygous carriers and patients. In both graphs means are indicated with a black line.
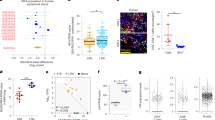
Supplementary Figure 4 DC immunophenotyping, HLA-DR expression in monocytes and DCs, Flt3 ligand production, and IL-12p40 secretion by whole blood.
a) Example of the gating strategy for Fig. 3a,d,g. b) Graphic representation of CD141+ cDC1s as the percentage of total PBMCs in healthy controls, healthy family members (heterozygous or homozygous carriers), patients with AR SPPL2a deficiency and patients with AD IRF8 deficiency from Fig. 3a. c) Levels of HLA-DR expression in monocytes, CD1c+ cells, cDC1s and pDCs from healthy controls and SPPL2a-deficient patients. Mean and S.E.M. are indicated. d) Flow cytometry histograms of the samples plotted in (c). Numbers indicate the MFI for HLA-DR PerCP-Cy5.5. e) CD1c+ cDCs (cDC2s) from controls and patients had higher levels of HLA-DR and CD11c expression than CD1c– cDCs. We assessed the presence of cDC2s in a CD1c-independent manner, by plotting CD11c vs. HLA-DR in live cells, FSC-A vs. SSC-A, FSC-A vs. FSC-W, HLA-DR+CD16– CD14–Lin-CD11c+CD1c+ or CD1c–. The CD1c– population in patients and controls did not display HLA-DRhigh CD11chigh expression, indicating that the smaller CD1c+ cDC2 population in the patients had not shifted to the CD1c– gate due to an absence of CD1c expression. These data provide further evidence of a lower levels of (cDC2s) CD1c+, HLA-DRhigh, and CD11chigh cDCs in the patients with AR SPPL2a deficiency or AD IRF8 deficiency. f) Flt3 ligand levels were determined in the plasma of healthy controls, patients with AR SPPL2a deficiency, patients with AD IRF8 deficiency, two heterozygous SPPL2A carriers (P1’s parents) and seven patients carrying heterozygous germline mutations of GATA2 causing monoMAC syndrome, as positive controls. g) IL-12p40 production by whole blood from healthy controls, WT and heterozygous family members and SPPL2a-deficient patients after stimulation with BCG or BCG+IFN-γ. In f and g mean and S.D. are indicated. The p values observed in this figure correspond to an unpaired two tailed t-test with a 95% confidence interval (* p < 0.05, ** p < 0.01).
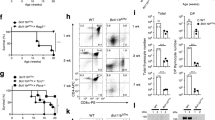
Supplementary Figure 5 Study of DC progenitors and in vitro differentiation.
a) Example of the gating strategy for the rest of the figure. b) Example of the gating strategy, starting from live single cells in (a), from a healthy control and a SPPL2a-deficient patient used for Fig. 4a. c) Graphic representation of total CD34+ progenitors in healthy controls and patients, obtained from (b) showing the mean and S.D. d) Gating strategy, starting from live single cells in (a), for a control and a SPPL2a-deficient patient for the graphs shown in Fig. 4b. e) Graphic representation of CD34+HLA-DR– DC progenitors obtained from (d) showing the mean and S.D. f) Gating strategy of early pre-DCs in healthy controls and SPPL2a-deficient patients, starting from the last gate in (a), the data for which are represented in Fig. 4c.
Supplementary Figure 6 Response to BCG and MTB in CD4+CCR6– T cells.
a) Representation of the percentage of IFN-γ+ cells in the T cell lines from the CCR6+ memory subsets upon stimulation with PMA+ionomycin. The dashed line separates samples analyzed in different experiments. b, c) IFN-γ secretion by T cell lines from CCR6+ memory subsets upon stimulation with PMA+ionomycin in (b) or activating anti-CD3 anti-CD28 Abs in (c). d) Representation of the percentage of IFN-γ+ cells in the T cell lines from CD4+CCR6– memory subsets upon stimulation with PMA+ionomycin. The dashed line separates samples analyzed in different experiments. e, f) IFN-γ secretion by T cell lines from CD4+CCR6– memory subsets upon stimulation with PMA+ionomycin (e) or anti CD3+anti-CD28 Abs (f). g) Frequency of CD4+ T cells reactive against the indicated antigens in the CCR6– memory subsets (containing Th1 and Th2 cells) from controls, patients with AR SPPL2a deficiency and patients with AD IRF8 deficiency. h) IFN-γ production by BCG- or i) MTB-specific CD4+ T cells from the CCR6– memory subsets. The vertical dashed line separates samples run in different experiments. An unpaired two-tailed t-test with a 95% confidence interval was performed (ns non-significative, **** p < 0.0001). j) Immunophenotyping for CCR4, CXCR3 and CCR6 of MTB- and flu-specific T cell clones from P1 and P2, showing that MTB-specific T cells are Th1* (CCR6+CXCR3+), whereas flu-specific T cells are Th1(CCR6–CXCR3+). k) MTB-specific T cell clones or flu-specific T cell clones from P1 and P2 were stimulated with MTB peptide pool and MTB lysate or flu vaccine, respectively. EBV-B cells from P1, non-transduced (NT), transduced with a retrovirus generated with an empty vector (EV) or a WT allele of SPPL2A (Fig. 2c) were used as APCs for these T cell clones. Proliferation of these T cell clones was measured by [3H]-thymidine assays and expressed as Δcpm. All graphs in this figure have the mean represented and from g to j also the S.E.M.
Supplementary Figure 7 Regulation of SPPL2a expression by IRF8 and gating for mouse experiments.
a) ChIP-Seq analysis of mouse bone marrow-derived macrophages and peritoneal macrophages left unstimulated, or stimulated with IFN-γ or TLR4 agonists (LPS or KLA). The Sppl2a promoter contains binding sites for STAT1, IRF1 and IRF8. RNA-seq profiles demonstrate an induction of Sppl2a expression following macrophage activation with IFN-γ. b) SPPL2a expression, as assessed by immunobloting (WB) and FACS in EBV-B cells from controls, one STAT1-deficient cell line, one IRF8-deficient cell line and one SPPL2a-deficient cell line. In the immunoblot, GAPDH was used as a loading control and the expression of IRF8, STAT1 and CD74 was also assessed. IRF8–/– denotes a cell line carrying the K108E mutation, which completely abolished the function of the protein but not its production (Hambleton et al. 2011). c) Example of the gating strategy used for (d). d) Graphical representation of SPPL2a expression in total lymphocytes, B cells and T cells in healthy controls and AD IRF8-deficient patients. e) Example of a representative FACS experiment gating used for the immunophenotyping graphs shown in Fig. 7 and in the rest of this figure. Hambleton, S. et al. IRF8 mutations and human dendritic-cell immunodeficiency. N Engl J Med 365, 127-138 (2011).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Tables 1–4
Rights and permissions
About this article
Cite this article
Kong, XF., Martinez-Barricarte, R., Kennedy, J. et al. Disruption of an antimycobacterial circuit between dendritic and helper T cells in human SPPL2a deficiency. Nat Immunol 19, 973–985 (2018). https://doi.org/10.1038/s41590-018-0178-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-018-0178-z
This article is cited by
-
Mendelian susceptibility to mycobacterial disease: an overview
Egyptian Journal of Medical Human Genetics (2023)
-
The transmembrane domain of Frey1 harbors a transplantable inhibitory motif for intramembrane proteases
Cellular and Molecular Life Sciences (2023)
-
Mendelian Susceptibility to Mycobacterial Disease (MSMD): Clinical, Immunological, and Genetic Features of 22 Patients from 15 Moroccan Kindreds
Journal of Clinical Immunology (2023)
-
Genetic Diagnosis of Inborn Errors of Immunity in an Emerging Country: a Retrospective Study of 216 Moroccan Patients
Journal of Clinical Immunology (2023)
-
Phagosomal signalling of the C-type lectin receptor Dectin-1 is terminated by intramembrane proteolysis
Nature Communications (2022)