Abstract
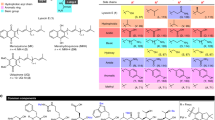
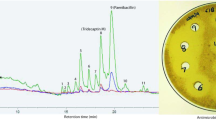
The rise of antibiotic resistance demands the acceleration of molecular diversification strategies to inspire new chemical entities for antibiotic medicines. We report here on the large-scale engineering of ribosomally synthesized and post-translationally modified antimicrobial peptides carrying the ring-forming amino acid lanthionine. New-to-nature variants featuring distinct properties were obtained by combinatorial shuffling of peptide modules derived from 12 natural antimicrobial lanthipeptides and processing by a promiscuous post-translational modification machinery. For experimental characterization, we developed the nanoFleming, a miniaturized and parallelized high-throughput inhibition assay. On the basis of a hit set of >100 molecules, we identified variants with improved activity against pathogenic bacteria and shifted activity profiles, and extrapolated design guidelines that will simplify the identification of peptide-based anti-infectives in the future.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The raw data from next generation sequencing of the peptide-encoding DNA libraries in Supplementary Fig. 2 are available in the NCBI Sequence Read Archive (SRA), accession number PRJNA511380.
Code availability
Data from next generation sequencing of the module libraries, from mass spectrometry and from MIC measurements were analyzed using scripts written in the programming language R. The code of the scripts is available from the corresponding authors upon reasonable request.
References
Newman, D. J. & Cragg, G. M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 79, 629–661 (2016).
Zengler, K. et al. Cultivating the uncultured. Proc. Natl Acad. Sci. USA 99, 15681–15686 (2002).
Baltz, R. H. Marcel Faber Roundtable: is our antibiotic pipeline unproductive because of starvation, constipation or lack of inspiration? J. Ind. Microbiol. Biotechnol. 33, 507–513 (2006).
Nichols, D. et al. Use of iChip for high-throughput in situ cultivation of ‘uncultivable’ microbial species. Appl. Environ. Microbiol. 76, 2445–2450 (2010).
Ling, L. L. et al. A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459 (2015).
Fischbach, M. A. & Walsh, C. T. Antibiotics for emerging pathogens. Science 325, 1089–1093 (2009).
Walsh, C. T. Insights into the chemical logic and enzymatic machinery of NRPS assembly lines. Nat. Prod. Rep. 33, 127–135 (2016).
Menzella, H. G. et al. Combinatorial polyketide biosynthesis by de novo design and rearrangement of modular polyketide synthase genes. Nat. Biotechnol. 23, 1171–1176 (2005).
Winn, M., Fyans, J. K., Zhuo, Y. & Micklefield, J. Recent advances in engineering nonribosomal peptide assembly lines. Nat. Prod. Rep. 33, 317–347 (2016).
Weissman, K. J. Genetic engineering of modular PKSs: from combinatorial biosynthesis to synthetic biology. Nat. Prod. Rep. 33, 203–230 (2016).
Arnison, P. G. et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160 (2013).
van Heel, A. J., Montalban-Lopez, M. & Kuipers, O. P. Evaluating the feasibility of lantibiotics as an alternative therapy against bacterial infections in humans. Expert Opin. Drug Metab. Toxicol 7, 675–680 (2011).
Legala, O. E., Yassi, H., Pflugmacher, S. & Neubauer, P. Pharmacological and pharmacokinetic properties of lanthipeptides undergoing clinical studies. Biotechnol. Lett. 39, 473–482 (2017).
Li, B. et al. Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine Cyanobacteria. Proc. Natl Acad. Sci. USA 107, 10430–10435 (2010).
Zhang, Q., Yang, X., Wang, H. & van der Donk, W. A. High divergence of the precursor peptides in combinatorial lanthipeptide biosynthesis. ACS Chem. Biol. 9, 2686–2694 (2014).
Oman, T. J. & van der Donk, W. A. Follow the leader: the use of leader peptides to guide natural product biosynthesis. Nat. Chem. Biol. 6, 9–18 (2010).
van Heel, A. J. et al. Discovery, production and modification of five novel lantibiotics using the promiscuous nisin modification machinery. ACS Synth. Biol. 5, 1146–1154 (2016).
Montalbán-López, M., van Heel, A. J. & Kuipers, O. P. Employing the promiscuity of lantibiotic biosynthetic machineries to produce novel antimicrobials. FEMS Microbiol. Rev. 41, 5–18 (2017).
Majchrzykiewicz, J. A. et al. Production of a class II two-component lantibiotic of Streptococcus pneumoniae using the class I nisin synthetic machinery and leader sequence. Antimicrob. Agents Chemother. 54, 1498–1505 (2010).
Dischinger, J., Basi Chipalu, S. & Bierbaum, G. Lantibiotics: promising candidates for future applications in health care. Int. J. Med. Microbiol. 304, 51–62 (2014).
Fleming, A. On the antibacterial action of cultures of a Penicillium, with special reference to their use in the isolation of B. influenzae. Br. J. Exp. Pathol. 10, 226–236 (1929).
Walser, M. et al. Novel method for high-throughput colony PCR screening in nanoliter-reactors. Nucleic Acids Res. 37, e57 (2009).
Walser, M., Leibundgut, R. M., Pellaux, R., Panke, S. & Held, M. Isolation of monoclonal microcarriers colonized by fluorescent E. coli. Cytom. Part A 73A, 788–798 (2008).
Meyer, A. et al. Optimization of a whole-cell biocatalyst by employing genetically encoded product sensors inside nanolitre reactors. Nat. Chem. 7, 673–678 (2015).
Roberts, T. M. et al. Identification and characterisation of a pH-stable GFP. Sci. Rep. 6, 28166 (2016).
Zhou, L., van Heel, A. J. & Kuipers, O. P. The length of a lantibiotic hinge region has profound influence on antimicrobial activity and host specificity. Front. Microbiol 6, 11 (2015).
Montalbán-López, M., Deng, J., van Heel, A. J. & Kuipers, O. P. Specificity and application of the lantibiotic protease NisP. Front. Microbiol 9, 160 (2018).
Rogers, A. Improved agar diffusion assay for nisin quantification. Food Biotechnol. 5, 161–168 (1991).
Khosa, S., Lagedroste, M. & Smits, S. H. J. Protein defense systems against the lantibiotic nisin: function of the immunity protein NisI and the resistance protein NSR. Front. Microbiol 7, 504 (2016).
Khosa, S. et al. Structural basis of lantibiotic recognition by the nisin resistance protein from Streptococcus agalactiae. Sci. Rep. 6, 18679 (2016).
Zhou, L., van Heel, A. J., Montalban-Lopez, M. & Kuipers, O. P. Potentiating the activity of nisin against Escherichia coli. Front. Cell Dev. Biol 4, 7 (2016).
Casini, A. et al. One-pot DNA construction for synthetic biology: the modular overlap-directed assembly with Linkers (MODAL) strategy. Nucleic Acids Res. 42, e7 (2014).
Kuipers, A. et al. NisT, the transporter of the lantibiotic nisin, can transport fully modified, dehydrated, and unmodified prenisin and fusions of the leader peptide with non-lantibiotic peptides. J. Biol. Chem. 279, 22176–22182 (2004).
Bernard, P. & Couturier, M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J. Mol. Biol. 226, 735–745 (1992).
van der Vossen, J. M., van der Lelie, D. & Venema, G. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl. Environ. Microbiol. 53, 2452–2457 (1987).
Dominy, C. N. & Andrews, D. W. Site-directed mutagenesis by inverse PCR. Methods Mol. Biol 235, 209–223 (2003).
Zheng, L. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32, e115–e115 (2004).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Schindelin, J., Rueden, C. T., Hiner, M. C. & Eliceiri, K. W. The ImageJ ecosystem: an open platform for biomedical image analysis. Mol. Reprod. Dev. 82, 518–529 (2015).
Green, M. R. & Sambrook, J. Molecular Cloning: A Laboratory Manual 4th edn (Cold Spring Harbor Laboratory Press, 2012).
Anthis, N. J. & Clore, G. M. Sequence-specific determination of protein and peptide concentrations by absorbance at 205 nm. Protein Sci. 22, 851–858 (2013).
Pelillo, M. et al. Calculation of the molar absorptivity of polyphenols by using liquid chromatography with diode array detection: the case of carnosic acid. J. Chromatogr. A 1023, 225–229 (2004).
Wiegand, I., Hilpert, K. & Hancock, R. E. W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175 (2008).
Gibb, S. & Strimmer, K. Maldiquant: a versatile R package for the analysis of mass spectrometry data. Bioinformatics 28, 2270–2271 (2012).
Acknowledgements
We thank A. van de Vries (Department of Biosystems Science and Engineering (D-BSSE), ETH Zürich) for assistance in protocol development, R. Pellaux, A. Meyer (both FGen GmbH), T. Roberts (D-BSSE) and A.J. van Heel (Department of Molecular Genetics, University of Groningen) for their valuable suggestions during the whole project. We thank the Genomics Facility Basel (C. Beisel, K. Eschbach, E.V. Burcklen, I. Nissen-Naidanow and M. Kohler, D-BSSE) for their help with next generation sequencing and S. Posada-Céspedes (D-BSSE) for her help with sequence analysis. Furthermore, the authors would like to thank A. Femmer (D-BSSE) for her excellent technical assistance during peptide characterization and thank S. Smits for the pNZ-SV-SaNSR construct. Last, S.S., M.M.-L., D.P., R.W., O.P.K. and S.P. would like to acknowledge funding from the ESF EUROCORES project ‘SYNMOD’ (grant number FP-017) and the EU FP7 project ‘SYNPEPTIDE’ (grant number 613981) and J.D. funding from the Chinese Scholarship Council (CSC).
Author information
Authors and Affiliations
Contributions
O.P.K. and S.P. conceived the study. D.P. and R.W. developed the DNA synthesis strategy. S.S., M.M.-L., D.P., R.W. and O.P.K. designed and prepared the combinatorial library. S.S. performed the library quality control and developed the nanoFleming assay, the idea for which had been conceived by M.H. O.P.K., M.M.-L. and S.S. designed and prepared the screening strains. S.S. did the screening, characterization and purification of the screening hits. S.S., M.M.-L. and J.D. determined the MICs of the peptides. R.W., M.H., O.P.K. and S.P. supervised the work. S.S., M.H., M.M.-L., O.P.K. and S.P. wrote the paper. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–11 and Supplementary Tables 1–7
Rights and permissions
About this article
Cite this article
Schmitt, S., Montalbán-López, M., Peterhoff, D. et al. Analysis of modular bioengineered antimicrobial lanthipeptides at nanoliter scale. Nat Chem Biol 15, 437–443 (2019). https://doi.org/10.1038/s41589-019-0250-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0250-5
This article is cited by
-
Genome mining unveils a class of ribosomal peptides with two amino termini
Nature Communications (2023)
-
Antiviral activities and applications of ribosomally synthesized and post-translationally modified peptides (RiPPs)
Cellular and Molecular Life Sciences (2021)
-
Minimal lactazole scaffold for in vitro thiopeptide bioengineering
Nature Communications (2020)
-
Features and applications of Ent35-MccV hybrid bacteriocin: current state and perspectives
Applied Microbiology and Biotechnology (2020)