Abstract
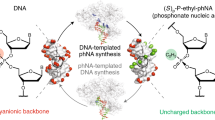
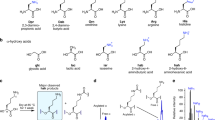
The chemical functionalities within biopolymers determine their physical properties and biological activities. The relationship between the side chains available to a biopolymer population and the potential functions of the resulting polymers, however, has proven difficult to study experimentally. Using seven sets of chemically diverse charged, polar, and nonpolar side chains, we performed cycles of artificial translation, in vitro selections for binding to either PCSK9 or IL-6 protein, and replication on libraries of random side chain-functionalized nucleic acid polymers. Polymer sequence convergence, bulk population target binding, affinity of individual polymers, and head-to-head competition among post-selection libraries collectively indicate that polymer libraries with nonpolar side chains outperformed libraries lacking these side chains. The presence of nonpolar groups, resembling functionality existing in proteins but missing from natural nucleic acids, thus may be strong determinants of binding activity. This factor may contribute to the apparent evolutionary advantage of proteins over their nucleic acid precursors for some molecular recognition tasks.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
High-throughput sequencing data will be available from the NCBI Sequence Read Archive under accession code SRP153119.
Change history
11 March 2019
In the version of this article originally published, several data points in Fig. 4c were shifted out of place during production. The corrected version of Fig. 4c is shown below. This error has been corrected in the PDF and HTML versions of the article.
References
Kamtekar, S., Schiffer, J. M., Xiong, H., Babik, J. M. & Hecht, M. H. Protein design by binary patterning of polar and nonpolar amino acids. Science 262, 1680–1685 (1993).
Davidson, A. R. & Sauer, R. T. Folded proteins occur frequently in libraries of random amino acid sequences. Proc. Natl Acad. Sci. USA 91, 2146–2150 (1994).
Heim, E. N. et al. Biologically active LIL proteins built with minimal chemical diversity. Proc. Natl Acad. Sci. USA 112, E4717–E4725 (2015).
Taylor, S. V., Walter, K. U., Kast, P. & Hilvert, D. Searching sequence space for protein catalysts. Proc. Natl Acad. Sci. USA 98, 10596–10601 (2001).
Beasley, J. R. & Hecht, M. H. Protein design: the choice of de novo sequences. J. Biol. Chem. 272, 2031–2034 (1997).
Reader, J. S. & Joyce, G. F. A ribozyme composed of only two different nucleotides. Nature 420, 841–844 (2002).
Ruff, K. M., Snyder, T. M. & Liu, D. R. Enhanced functional potential of nucleic acid aptamer libraries patterned to increase secondary structure. J. Am. Chem. Soc. 132, 9453–9464 (2010).
Rogers, J. & Joyce, G. F. A ribozyme that lacks cytidine. Nature 402, 323–325 (1999).
Schlosser, K. & Li, Y. DNAzyme-mediated catalysis with only guanosine and cytidine nucleotides. Nucleic Acids Res. 37, 413–420 (2009).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Ellington, A. D. & Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
Robertson, D. L. & Joyce, G. F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 344, 467–468 (1990).
Bock, L. C., Griffin, L. C., Latham, J. A., Vermaas, E. H. & Toole, J. J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 355, 564–566 (1992).
Ellington, A. D. & Szostak, J. W. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 355, 850–852 (1992).
Keefe, A. D., Pai, S. & Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug. Discov. 9, 537–550 (2010).
Breaker, R. R. & Joyce, G. F. A DNA enzyme that cleaves RNA. Chem. Biol. 1, 223–229 (1994).
Li, Y., Liu, Y. & Breaker, R. R. Capping DNA with DNA. Biochemistry 39, 3106–3114 (2000).
Silverman, S. K. In vitro selection, characterization, and application of deoxyribozymes that cleave RNA. Nucleic Acids Res. 33, 6151–6163 (2005).
Schlosser, K. & Li, Y. Biologically inspired synthetic enzymes made from DNA. Chem. Biol. 16, 311–322 (2009).
Walsh, S. M., Sachdeva, A. & Silverman, S. K. DNA catalysts with tyrosine kinase activity. J. Am. Chem. Soc. 135, 14928–14931 (2013).
Silverman, S. K. Catalytic DNA: scope, applications, and biochemistry of deoxyribozymes. Trends. Biochem. Sci. 41, 595–609 (2016).
Gold, L. et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One 5, e15004–e15017 (2010).
Ren, X., Gelinas, A. D., von Carlowitz, I., Janjic, N. & Pyle, A. M. Structural basis for IL-1α recognition by a modified DNA aptamer that specifically inhibits IL-1α signaling. Nat. Commun. 8, 810 (2017).
Ochsner, U. A., Katilius, E. & Janjic, N. Detection of Clostridium difficile toxins A, B and binary toxin with slow off-rate modified aptamers. Diagn. Microbiol. Infect. Dis. 76, 278–285 (2013).
Zhou, C. et al. DNA-catalyzed amide hydrolysis. J. Am. Chem. Soc. 138, 2106–2109 (2016).
Hollenstein, M., Hipolito, C. J., Lam, C. H. & Perrin, D. M. A DNAzyme with three protein-like functional groups: enhancing catalytic efficiency of M2+-independent RNA cleavage. Chembiochem 10, 1988–1992 (2009).
Sidorov, A. V., Grasby, J. A. & Williams, D. M. Sequence-specific cleavage of RNA in the absence of divalent metal ions by a DNAzyme incorporating imidazolyl and amino functionalities. Nucleic Acids Res. 32, 1591–1601 (2004).
Santoro, S. W., Joyce, G. F., Sakthivel, K., Gramatikova, S. & Barbas, C. F. III. RNA cleavage by a DNA enzyme with extended chemical functionality. J. Am. Chem. Soc. 122, 2433–2439 (2000).
Thomas, J. M., Yoon, J.-K. & Perrin, D. M. Investigation of the catalytic mechanism of a synthetic DNAzyme with protein-like functionality: an RNaseA mimic? J. Am. Chem. Soc. 131, 5648–5658 (2009).
Rohloff, J. C. et al. Nucleic acid ligands with protein-like side chains: modified aptamers and their use as diagnostic and therapeutic agents. Mol. Ther. Nucleic Acids 3, e201 (2014).
Gawande, B. N. et al. Selection of DNA aptamers with two modified bases. Proc. Natl Acad. Sci. USA 114, 2898–2903 (2017).
Davies, D. R. et al. Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets. Proc. Natl Acad. Sci. USA 109, 19971–19976 (2012).
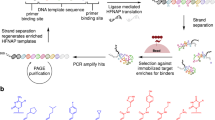
Hili, R., Niu, J. & Liu, D. R. DNA ligase-mediated translation of DNA into densely functionalized nucleic acid polymers. J. Am. Chem. Soc. 135, 98–101 (2013).
Chen, Z., Lichtor, P. A., Berliner, A. P., Chen, J. C. & Liu, D. R. Evolution of sequence-defined highly functionalized nucleic acid polymers. Nat. Chem. 10, 420–427 (2018).
Kong, D., Lei, Y., Yeung, W. & Hili, R. Enzymatic synthesis of sequence-defined synthetic nucleic acid polymers with diverse functional groups. Angew. Chem. Int. Ed. Engl. 55, 13164–13168 (2016).
Kong, D., Yeung, W. & Hili, R. In vitro selection of diversely functionalized aptamers. J. Am. Chem. Soc. 139, 13977–13980 (2017).
Lei, Y., Washington, J. & Hili, R. Efficiency and fidelity of T3 DNA ligase in ligase-catalysed oligonucleotide polymerisations. Org. Biomol. Chem. 1, 0076 (2018).
Rose, G. D., Geselowitz, A. R., Lesser, G. J., Lee, R. H. & Zehfus, M. H. Hydrophobicity of amino acid residues in globular proteins. Science 229, 834–838 (1985).
Koide, S. & Sidhu, S. S. The importance of being tyrosine: lessons in molecular recognition from minimalist synthetic binding proteins. ACS. Chem. Biol. 4, 325–334 (2009).
Ramaraj, T., Angel, T., Dratz, E. A., Jesaitis, A. J. & Mumey, B. Antigen-antibody interface properties: composition, residue interactions, and features of 53 non-redundant structures. Biochim. Biophys. Acta 1824, 520–532 (2012).
Hunter, C. A. & Jones, S. A. IL-6 as a keystone cytokine in health and disease. Nat. Immunol. 16, 448–457 (2015).
Halford, B. Cholesterol-lowering PCSK9 inhibitors near market entry. Chem. Eng. News 93, 10–14 (2015).
Sabatine, M. S. et al. Evolocumab and clinical outcomes in patients with cardiovascular disease. N. Engl. J. Med. 376, 1713–1722 (2017).
Gupta, S. et al. Chemically modified DNA aptamers bind interleukin-6 with high affinity and inhibit signaling by blocking its interaction with interleukin-6 receptor. J. Biol. Chem. 289, 8706–8719 (2014).
Gelinas, A. D. et al. Crystal structure of interleukin-6 in complex with a modified nucleic acid ligand. J. Biol. Chem. 289, 8720–8734 (2014).
Zimmermann, B., Gesell, T., Chen, D., Lorenz, C. & Schroeder, R. Monitoring genomic sequences during SELEX using high-throughput sequencing: neutral SELEX. PLoS One 5, e9169 (2010).
Lipinski, C. A., Lombardo, F., Dominy, B. W. & Feeney, P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 23, 3–25 (1997)..
Hughes, J. D. et al. Physiochemical drug properties associated with in vivo toxicological outcomes. Bioorg. Med. Chem. Lett. 18, 4872–4875 (2008).
Acknowledgements
This work was supported by DARPA N66001-14-2-4053, NIH R35 GM118062, and the Howard Hughes Medical Institute. Z.C. was partially supported by the Y. Kishi Graduate Prize in Chemistry and Chemical Biology sponsored by the Eisai Corporation. The authors thank A. Chan, J.P. Maianti, M. Packer, D.L. Usanov, B. Thuronyi, and C. Wilson for helpful comments and discussions. We also thank K. Arnett and R. Stoller for guidance with SPR experiments and analysis.
Author information
Authors and Affiliations
Contributions
P.A.L., Z.C., J.C.C. performed experiments; P.A.L. and N.H.E. developed the SPR methodology; P.A.L., Z.C., and D.R.L. designed the research and wrote the manuscript; all authors reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
Z.C., P.A.L., and D.R.L. have filed patent applications on DNA-templated polymerization.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–3, Supplementary Figures 1–12, and Supplementary Notes 1–4
Rights and permissions
About this article
Cite this article
Lichtor, P.A., Chen, Z., Elowe, N.H. et al. Side chain determinants of biopolymer function during selection and replication. Nat Chem Biol 15, 419–426 (2019). https://doi.org/10.1038/s41589-019-0229-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0229-2
This article is cited by
-
Reconstruction of evolving gene variants and fitness from short sequencing reads
Nature Chemical Biology (2021)
-
Modified nucleic acids: replication, evolution, and next-generation therapeutics
BMC Biology (2020)
-
ATP-powered molecular recognition to engineer transient multivalency and self-sorting 4D hierarchical systems
Nature Communications (2020)