Abstract
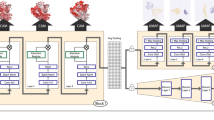
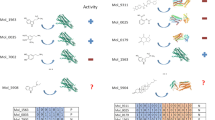
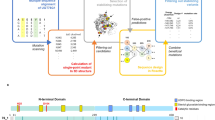
The elucidation and prediction of how changes in a protein result in altered activities and selectivities remain a major challenge in chemistry. Two hurdles have prevented accurate family-wide models: obtaining (i) diverse datasets and (ii) suitable parameter frameworks that encapsulate activities in large sets. Here, we show that a relatively small but broad activity dataset is sufficient to train algorithms for functional prediction over the entire glycosyltransferase superfamily 1 (GT1) of the plant Arabidopsis thaliana. Whereas sequence analysis alone failed for GT1 substrate utilization patterns, our chemical–bioinformatic model, GT-Predict, succeeded by coupling physicochemical features with isozyme-recognition patterns over the family. GT-Predict identified GT1 biocatalysts for novel substrates and enabled functional annotation of uncharacterized GT1s. Finally, analyses of GT-Predict decision pathways revealed structural modulators of substrate recognition, thus providing information on mechanisms. This multifaceted approach to enzyme prediction may guide the streamlined utilization (and design) of biocatalysts and the discovery of other family-wide protein functions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Activity datasets, mass spectrograms, and the protein FASTA sequences used herein are included in a package available through the Oxford University Research Archive at https://doi.org/10.5287/bodleian:zg5195kaE.
References
Todd, A. E., Orengo, C. A. & Thornton, J. M. Evolution of function in protein superfamilies, from a structural perspective. J. Mol. Biol. 307, 1113–1143 (2001).
Gerlt, J. A. & Babbitt, P. C. Mechanistically diverse enzyme superfamilies: the importance of chemistry in the evolution of catalysis. Curr. Opin. Chem. Biol. 2, 607–612 (1998).
Friedmann, D. R. & Marmorstein, R. Structure and mechanism of non-histone protein acetyltransferase enzymes. FEBS J. 280, 5570–5581 (2013).
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M. & Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 (2014).
Li, T. et al. Characterization and prediction of lysine (K)-acetyl-transferase specific acetylation sites. Mol. Cell. Proteomics. 11, M111.011080 (2012).
Lim, E.-K. et al. Evolution of substrate recognition across a multigene family of glycosyltransferases in Arabidopsis. Glycobiology 13, 139–145 (2003).
Modolo, L. V. et al. A functional genomics approach to (iso)flavonoid glycosylation in the model legume Medicago truncatula. Plant Mol. Biol. 64, 499–518 (2007).
Lairson, L. L., Henrissat, B., Davies, G. J. & Withers, S. G. Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555 (2008).
Cartwright, A. M., Lim, E.-K., Kleanthous, C. & Bowles, D. J. A kinetic analysis of regiospecific glucosylation by two glycosyltransferases of Arabidopsis thaliana: domain swapping to introduce new activities. J. Biol. Chem. 283, 15724–15731 (2008).
Todd, A. E., Orengo, C. A. & Thornton, J. M. Plasticity of enzyme active sites. Trends. Biochem. Sci. 27, 419–426 (2002).
Gloster, T. M. Advances in understanding glycosyltransferases from a structural perspective. Curr. Opin. Struct. Biol. 28, 131–141 (2014).
Harper, K. C. & Sigman, M. S. Predicting and optimizing asymmetric catalyst performance using the principles of experimental design and steric parameters. Proc. Natl. Acad. Sci. USA 108, 2179–2183 (2011).
Kries, H., Blomberg, R. & Hilvert, D. De novo enzymes by computational design. Curr. Opin. Chem. Biol. 17, 221–228 (2013).
Yang, M., Brazier, M., Edwards, R. & Davis, B. G. High-throughput mass-spectrometry monitoring for multisubstrate enzymes: determining the kinetic parameters and catalytic activities of glycosyltransferases. Chembiochem 6, 346–357 (2005).
Flint, J. et al. Structural dissection and high-throughput screening of mannosylglycerate synthase. Nat. Struct. Mol. Biol. 12, 608–614 (2005).
Yang, M., Davies, G. J. & Davis, B. G. A glycosynthase catalyst for the synthesis of flavonoid glycosides. Angew. Chem. Int. Edn Engl. 46, 3885–3888 (2007).
Backus, K. M. et al. Uptake of unnatural trehalose analogs as a reporter for Mycobacterium tuberculosis. Nat. Chem. Biol. 7, 228–235 (2011).
Offen, W. et al. Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 25, 1396–1405 (2006).
Brazier-Hicks, M. et al. Characterization and engineering of the bifunctional N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc. Natl. Acad. Sci. USA 104, 20238–20243 (2007).
McLeod, M. C. et al. Probing chemical space with alkaloid-inspired libraries. Nat. Chem. 6, 133–140 (2014).
Li, Y., Baldauf, S., Lim, E. K. & Bowles, D. J. Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J. Biol. Chem. 276, 4338–4343 (2001).
Breiman, L., Friedman, J. H., Olshen, R. A. & Stone, C. J. Classification and Regression Trees (Wadsworth & Brooks, Monterey, CA, 1984).
Kotera, M., Goto, S. & Kanehisa, M. Predictive genomic and metabolomic analysis for the standardization of enzyme data. Perspect. Sci. 1, 24–32 (2014).
Sánchez-Rodríguez, A. et al. A network-based approach to identify substrate classes of bacterial glycosyltransferases. BMC Genomics 15, 349 (2014).
Smith, T. F. & Waterman, M. S. Identification of common molecular subsequences. J. Mol. Biol. 147, 195–197 (1981).
Shao, H. et al. Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell 17, 3141–3154 (2005).
Modolo, L. V. et al. Crystal structures of glycosyltransferase UGT78G1 reveal the molecular basis for glycosylation and deglycosylation of (iso)flavonoids. J. Mol. Biol. 392, 1292–1302 (2009).
Yang, M. et al. Probing the breadth of macrolide glycosyltransferases: in vitro remodeling of a polyketide antibiotic creates active bacterial uptake and enhances potency. J. Am. Chem. Soc. 127, 9336–9337 (2005).
Venturelli, S. et al. Resveratrol as a pan-HDAC inhibitor alters the acetylation status of histone [corrected] proteins in human-derived hepatoblastoma cells. PLoS ONE 8, e73097 (2013).
Kjaer, T. N. et al. Resveratrol reduces the levels of circulating androgen precursors but has no effect on, testosterone, dihydrotestosterone, PSA levels or prostate volume: a 4-month randomised trial in middle-aged men. Prostate 75, 1255–1263 (2015).
Turner, R. S. et al. A randomized, double-blind, placebo-controlled trial of resveratrol for Alzheimer disease. Neurology 85, 1383–1391 (2015).
Tomé-Carneiro, J. et al. Resveratrol and clinical trials: the crossroad from in vitro studies to human evidence. Curr. Pharm. Des. 19, 6064–6093 (2013).
Pandey, R. P. et al. Enzymatic biosynthesis of novel resveratrol glucoside and glycoside derivatives. Appl. Environ. Microbiol. 80, 7235–7243 (2014).
Weis, M., Lim, E.-K., Bruce, N. & Bowles, D. Regioselective glucosylation of aromatic compounds: screening of a recombinant glycosyltransferase library to identify biocatalysts. Angew. Chem. Int. Ed. Engl. 45, 3534–3538 (2006).
Burns, J., Yokota, T., Ashihara, H., Lean, M. E. & Crozier, A. Plant foods and herbal sources of resveratrol. J. Agric. Food. Chem. 50, 3337–3340 (2002).
Heide, L. The aminocoumarins: biosynthesis and biology. Nat. Prod. Rep. 26, 1241–1250 (2009).
Peneff, C. et al. Crystal structures of two human pyrophosphorylase isoforms in complexes with UDPGlc(Gal)NAc: role of the alternatively spliced insert in the enzyme oligomeric assembly and active site architecture. EMBO J. 20, 6191–6202 (2001).
Unligil, U. M. et al. X-ray crystal structure of rabbit N-acetylglucosaminyltransferase I: catalytic mechanism and a new protein superfamily. EMBO J. 19, 5269–5280 (2000).
Pearson, W. R. Protein function prediction: problems and pitfalls. Curr. Protoc. Bioinformatics 51, 12.1 –4.12.8 (2015).
Tyagi, S. & Pleiss, J. Biochemical profiling in silico: predicting substrate specificities of large enzyme families. J. Biotechnol. 124, 108–116 (2006).
Zhao, S. et al. Discovery of new enzymes and metabolic pathways by using structure and genome context. Nature 502, 698–702 (2013).
Nembri, S., Grisoni, F., Consonni, V. & Todeschini, R. In silico prediction of cytochrome P450-drug interaction: QSARs for CYP3A4 and CYP2C9. Int. J. Mol. Sci. 17, E914 (2016).
Dong, D., Ako, R., Hu, M. & Wu, B. Understanding substrate selectivity of human UDP-glucuronosyltransferases through QSAR modeling and analysis of homologous enzymes. Xenobiotica 42, 808–820 (2012).
Wang, T., Yuan, X. S., Wu, M.-B., Lin, J.-P. & Yang, L.-R. The advancement of multidimensional QSAR for novel drug discovery: where are we headed? Expert Opin. Drug Discov. 12, 769–784 (2017).
Law, V. et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 42, D1091–D1097 (2014).
Udayakumar, M. et al. PMDB: plant metabolome database: a metabolomic approach. Med. Chem. Res. 21, 47–52 (2012).
Schmid, J., Heider, D., Wendel, N. J., Sperl, N. & Sieber, V. Bacterial glycosyltransferases: challenges and opportunities of a highly diverse enzyme class toward tailoring natural products. Front. Microbiol. 7, 182 (2016).
Osmani, S. A., Bak, S. & Møller, B. L. Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry 70, 325–347 (2009).
Davies, G. J., Planas, A. & Rovira, C. Conformational analyses of the reaction coordinate of glycosidases. Acc. Chem. Res. 45, 308–316 (2012).
Newton, M. S. et al. Structural and functional innovations in the real-time evolution of new (βα)8 barrel enzymes. Proc. Natl. Acad. Sci. USA 114, 4727–4732 (2017).
Roy, A., Kucukural, A. & Zhang, Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010).
Mackenzie, P. I. et al. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet. Genomics 15, 677–685 (2005).
Lim, E.-K. et al. Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in Arabidopsis. J. Biol. Chem. 276, 4344–4349 (2001).
Berthold, M. R. et al. in Data Anal., Mach. Learn.Appl.: Proc. 31st Annu. Conf. Gesellschaft für Klassifikation e.V., Albert-Ludwigs-Universität Freiburg, March 7–9, 2007 (eds. Preisach, C. et al.) 319–326 (Springer, Berlin, 2008).
Sauer, W. H. B. & Schwarz, M. K. Molecular shape diversity of combinatorial libraries: a prerequisite for broad bioactivity. J. Chem. Inf. Comput. Sci. 43, 987–1003 (2003).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 (1997).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Rice, P., Longden, I. & Bleasby, A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277 (2000).
Johnson, S. C. Hierarchical clustering schemes. Psychometrika 32, 241–254 (1967).
Kohavi, R. A study of cross-validation and bootstrap for accuracy estimation and model selection. in IJCAI’95 Proc. 14th Int. Joint Conf. Artif. Intel. Vol. 2, 1137–1143 (Morgan Kaufmann, San Francisco, 1995).
Matthews, B. W. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta 405, 442–451 (1975).
Pearson, W. R. Selecting the right similarity-scoring matrix. Curr. Protoc. Bioinformatics 43, 5.1–3.5.9 (2013).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Learmonth, D. A. Novel convenient synthesis of the 3‐O‐β‐D‐ and 4′‐O β‐D‐glucopyranosides of trans‐resveratrol. Synth. Commun. 34, 1565–1575 (2004).
Acknowledgements
We gratefully acknowledge A. Osbourne (JIC) for contribution of A. strigosa GT1 genes As08 (UGT74H5) and As09 (UGT88C4); R. Edwards and M. Brazier-Hicks for sharing activity data; and I. Mear for assistance with coding. This work was funded by the BBSRC (EGA16205, EGA16206, and EGA17763) and the EPSRC (the UK Catalysis Hub, EP/K014668/1 and EP/M013219/1).
Author information
Authors and Affiliations
Contributions
G.J.D., D.J.B., M.G.D., S.J.R., and B.G.D. designed the research; M.Y., C.F., and K.V.L. performed the research; M.Y., C.F., K.V.L., E.-K.L. W.A.O., G.J.D., S.J.R., and B.G.D. analyzed the data; G.J.D., D.J.B., M.G.D., S.J.R., and B.G.D. wrote the paper; all authors read and commented on the paper. M.Y. and C.F. contributed equally to this work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–9, Supplementary Figures 1–16, and Supplementary Note
Supplementary Video 1
Installation and use of GT-Predict on Microsoft Windows 10
Rights and permissions
About this article
Cite this article
Yang, M., Fehl, C., Lees, K.V. et al. Functional and informatics analysis enables glycosyltransferase activity prediction. Nat Chem Biol 14, 1109–1117 (2018). https://doi.org/10.1038/s41589-018-0154-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-018-0154-9
This article is cited by
-
A general model to predict small molecule substrates of enzymes based on machine and deep learning
Nature Communications (2023)
-
Reconstitution of monoterpene indole alkaloid biosynthesis in genome engineered Nicotiana benthamiana
Communications Biology (2022)
-
Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides
Nature Communications (2021)
-
Divining sugar substrates
Nature Chemical Biology (2018)