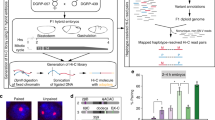
Abstract
Chromatin topology is intricately linked to gene expression, yet its functional requirement remains unclear. Here, we comprehensively assessed the interplay between genome topology and gene expression using highly rearranged chromosomes (balancers) spanning ~75% of the Drosophila genome. Using transheterozyte (balancer/wild-type) embryos, we measured allele-specific changes in topology and gene expression in cis, while minimizing trans effects. Through genome sequencing, we resolved eight large nested inversions, smaller inversions, duplications and thousands of deletions. These extensive rearrangements caused many changes to chromatin topology, disrupting long-range loops, topologically associating domains (TADs) and promoter interactions, yet these are not predictive of changes in expression. Gene expression is generally not altered around inversion breakpoints, indicating that mis-appropriate enhancer–promoter activation is a rare event. Similarly, shuffling or fusing TADs, changing intra-TAD connections and disrupting long-range inter-TAD loops does not alter expression for the majority of genes. Our results suggest that properties other than chromatin topology ensure productive enhancer–promoter interactions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All raw data, which consist of 75 demultiplexed files, were submitted to ArrayExpress (https://www.ebi.ac.uk/arrayexpress/browse.html) under accession numbers E-MTAB-7510 (whole-genome and mate-pair sequencing), E-MTAB-7512 (Hi-C), E-MTAB-7513 (Capture-C) and E-MTAB-7511 (RNA-Seq). The Hi-C contact maps, RNA-Seq read counts and other processed data are available on the Furlong Laboratory web page at http://furlonglab.embl.de/data.
Code availability
Custom code used for the analysis is available at https://github.com/ajank/balancer-paper.
References
Bulger, M. & Groudine, M. Functional and mechanistic diversity of distal transcription enhancers. Cell 144, 327–339 (2011).
Levine, M. Transcriptional enhancers in animal development and evolution. Curr. Biol. 20, R754–R763 (2010).
Spitz, F. & Furlong, E. E. M. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 13, 613–626 (2012).
Furlong, E. E. M. & Levine, M. Developmental enhancers and chromosome topology. Science 361, 1341–1345 (2018).
Jin, F. et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503, 290–294 (2013).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Nora, E. P. et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012).
Rao, S. S. P. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Sexton, T. et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472 (2012).
Lupiáñez, D. G. et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012–1025 (2015).
Franke, M. et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538, 265–269 (2016).
Tsujimura, T. et al. A discrete transition zone organizes the topological and regulatory autonomy of the adjacent Tfap2c and Bmp7 genes. PLoS Genet. 11, e1004897 (2015).
Narendra, V. et al. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347, 1017–1021 (2015).
Guo, Y. et al. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162, 900–910 (2015).
Lettice, L. A. et al. Enhancer-adoption as a mechanism of human developmental disease. Hum. Mutat. 32, 1492–1499 (2011).
Northcott, P. A. et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 511, 428–434 (2014).
Flavahan, W. A. et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114 (2016).
Hnisz, D. et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351, 1454–1458 (2016).
Northcott, P. A. et al. The whole-genome landscape of medulloblastoma subtypes. Nature 547, 311–317 (2017).
Weischenfeldt, J. et al. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat. Genet. 49, 65–74 (2017).
Shen, Y. et al. Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome. Science 355, eaaf4791 (2017).
Shao, Y. et al. Creating a functional single-chromosome yeast. Nature 560, 331–335 (2018).
Lee, H. et al. Effects of gene dose, chromatin, and network topology on expression in Drosophila melanogaster. PLoS Genet. 12, e1006295 (2016).
Meadows, L. A., Chan, Y. S., Roote, J. & Russell, S. Neighbourhood continuity is not required for correct testis gene expression in Drosophila. PLoS Biol. 8, e1000552 (2010).
Rodríguez-Carballo, E. et al. The HoxD cluster is a dynamic and resilient TAD boundary controlling the segregation of antagonistic regulatory landscapes. Genes Dev. 31, 2264–2281 (2017).
Nora, E. P. et al. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930–944.e22 (2017).
Splinter, E. et al. CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes Dev. 20, 2349–2354 (2006).
Rao, S. S. P. et al. Cohesin loss eliminates all loop domains. Cell 171, 305–320.e24 (2017).
Schwarzer, W. et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56 (2017).
Wutz, G. et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 36, 3573–3599 (2017).
Oster, I. I. A new crossing-over suppressor in chromosome 2 effective in the presence of heterologous inversions. Drosophila Inform. Serv. 30, 145 (1956).
Tinderholt, V. New mutants report. Drosophila Inform. Serv. 34, 53–54 (1960).
Ashburner, M., Golic, K. G. & Hawley, R. S. Drosophila: A Laboratory Handbook (Cold Spring Harbor Laboratory Press, 2005).
Korbel, J. O. et al. Paired-end mapping reveals extensive structural variation in the human genome. Science 318, 420–426 (2007).
Weischenfeldt, J., Symmons, O., Spitz, F. & Korbel, J. O. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat. Rev. Genet. 14, 125–138 (2013).
Mackay, T. F. C. et al. The Drosophila melanogaster genetic reference panel. Nature 482, 173–178 (2012).
Zichner, T. et al. Impact of genomic structural variation in Drosophila melanogaster based on population-scale sequencing. Genome Res. 23, 568–579 (2013).
Lindsley, D. L. & Zimm, G. G. The Genome of Drosophila Melanogaster (Academic Press, 1992).
Sudmant, P. H. et al. An integrated map of structural variation in 2,504 human genomes. Nature 526, 75–81 (2015).
Miller, D. E. et al. The molecular and genetic characterization of second chromosome balancers in Drosophila melanogaster. G3 (Bethesda) 8, 1161–1171 (2018).
Miller, D. E., Cook, K. R., Arvanitakis, A. V. & Hawley, R. S. Third chromosome balancer inversions disrupt protein-coding genes and influence distal recombination events in Drosophila melanogaster. G3 (Bethesda) 6, 1959–1967 (2016).
Huang, W. et al. Natural variation in genome architecture among 205 Drosophila melanogaster genetic reference panel lines. Genome Res. 24, 1193–1208 (2014).
Thomas, S. et al. Dynamic reprogramming of chromatin accessibility during Drosophila embryo development. Genome Biol. 12, R43 (2011).
Ramírez, F. et al. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun. 9, 189 (2018).
Ogiyama, Y., Schuettengruber, B., Papadopoulos, G. L., Chang, J.-M. & Cavalli, G. Polycomb-dependent chromatin looping contributes to gene silencing during Drosophila development. Mol. Cell 71, 73–88.e5 (2018).
Spielmann, M., Lupiáñez, D. G. & Mundlos, S. Structural variation in the 3D genome. Nat. Rev. Genet. 19, 453–467 (2018).
Mardin, B. R. et al. A cell‐based model system links chromothripsis with hyperploidy. Mol. Syst. Biol. 11, 828 (2015).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997 (2013).
Garrison, E. & Marth, G. Haplotype-based variant detection from short-read sequencing. Preprint at https://arxiv.org/abs/1207.3907 (2012).
Tan, A., Abecasis, G. R. & Kang, H. M. Unified representation of genetic variants. Bioinformatics 31, 2202–2204 (2015).
Rausch, T. et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28, i333–i339 (2012).
Untergasser, A. et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 40, e115 (2012).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 15, 550 (2014).
Strimmer, K. fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics 24, 1461–1462 (2008).
Bonn, S. et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 44, 148–156 (2012).
Davies, J. O. J. et al. Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat. Methods 13, 74–80 (2016).
Zhao, H. et al. CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics 30, 1006–1007 (2014).
Cairns, J. et al. CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome Biol. 17, 127 (2016).
Loader, C. locfit: Local Regression, Likelihood and Density Estimation https://CRAN.R-project.org/package=locfit (2013).
Cusanovich, D. A. et al. The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature 555, 538–542 (2018).
Zinzen, R. P., Girardot, C., Gagneur, J., Braun, M. & Furlong, E. E. M. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 462, 65–70 (2009).
Acknowledgements
We thank all members of the Furlong Laboratory for discussions and comments on the manuscript. We thank M. Davis, H. Gustafson, D. Garfield, T. Rausch and S. Waszak for useful discussions and suggestions at the various stages of the project. This work was technically supported by the EMBL Genomics Core Facility, with specific thanks to R. Hercog for WGS library preparation. This work was financially supported by an FRM grant (AJE20161236686) to Y.G.-H., the EMBL International PhD Programme to S.M., an EU Horizon 2020 Marie Skłodowska-Curie grant (708111) to A.J., an ERC starting grant (336045) to J.O.K. and ERC advanced grant DeCRyPT (787611) to E.E.M.F.
Author information
Authors and Affiliations
Contributions
Y.G.-H. and E.E.M.F. designed the study. Y.G.-H., A.J., S.M., J.O.K. and E.E.M.F. analyzed the results. Y.G.-H., A.J., S.M. and E.E.M.F. wrote the manuscript. Y.G.-H. performed all of the experiments, except the mate-pair library experiment, which was performed by R.R.V. S.M. performed the SNV and structural variant calling and RNA-Seq analysis. A.J. performed the Hi-C and Capture-C data analysis. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Crossing scheme to generate heterozygote embryos.
a, Males from a double balancer line (If/CyO; Sb/TM3) were crossed to females from an isogenic wild-type “virginizer” line (+/+; +/+; see Methods). Resulting heterozygous males from the F1 generation (+/CyO; +/TM3) were back-crossed to females of the parental line. Embryos from the resulting N1pat generation (pool of 4 genotypes) were collected to perform allele-specific RNA-seq, Hi-C and Capture-C experiments. The reciprocal N1mat generation, used for RNA-seq, was obtained by back-crossing heterozygous females from the F1 generation to the male parental line. b, Table listing all datasets generated in the study, including genotype and developmental stage.
Supplementary Figure 2 Example and characteristics of Structural Variants in the balancer chromosomes.
a, A 258 kb balancer-specific duplication in chr2L. Top to bottom: wild-type and balancer Hi-C contact maps, log2 fold change before normalization (balancer/wild-type, red/blue), gene models, and differential gene expression (balancer/wild-type, log2 fold change). Differentially expressed genes (DE-genes) in orange, non-differentially expressed (non-DE) genes in blue and non-tested genes (lowly expressed or lacking SNVs) in grey. Hi-C contacts (~two-fold increase in balancer) confirm the duplication, inserted in an inverted orientation shown in (b). Expressed genes have a ~2-fold increase in allelic expression, as expected. Dotted lines indicate TAD boundaries. b, Model of the duplication and associated changes in TAD structure. Arrows mark the duplicated fragment, gradient indicates newly created contacts. c, Number of DNase hypersensitive (DHS) peaks deleted by ≥ 5% of their length in the wild-type (top) and balancer (bottom), indicated by orange vertical line. Deletions generally overlap a DHS less frequently than expected from randomly shuffling deleted regions in the genome (blue, green bars, respectively), as anticipated due to purifying selection. d-e, A 17.5 kb balancer-specific deletion at the breakpoint of the inversion chr3R:20.32 Mb: Wild-type (d) and balancer (e) Hi-C contact maps, deleted region in balancer highlighted in grey (d). DHS, gene models, and differential gene expression (balancer/wild-type, log2 fold change) are shown underneath. DE-genes in orange, non-DE genes in blue and non-tested genes (lowly expressed or lacking SNVs) in grey. Dashed purple lines indicate the location of the inversion boundary.
Supplementary Figure 3 Changes in gene expression between balancer and wild-type haplotypes.
a, Change in gene expression (balancer/wild-type, log2 fold change) in F1 heterozygous and single balancer (CyO only or TM3 only) adult fly heads (mixed sexes). Genes with significant (two-sided Wald test, 5% FDR) effect of genotype on differential expression (DE-genes) highlighted in blue, Pearson correlation coefficient (r) indicated. b, Same as (a), but comparing F1 heterozygous adult female heads to mixed sex heads as a proxy for expected biological noise. c, Change in allele-specific gene expression (log2 fold change) for testable genes with exons overlapping an allele-specific deletion or duplication. Fully duplicated genes are shown separately in Fig. 2b. Points, individual genes (180 genes partially deleted in balancer, 116 partially deleted in wild-type, 13 partially duplicated in balancer, 14 partially duplicated in wild-type); center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; dashed lines, two-fold increase or decrease.
Supplementary Figure 4 Extensive genome rearrangements within TADs have limited impact on gene expression.
a, Insulation score profiles, calculated at 21,800 genomic bins, are highly correlated between wild-type and balancer haplotypes when comparing replicates (R1 and R2) for each allele, even when considering regions affected by nested inversion breakpoints (red). Pearson correlation coefficient (r) indicated. Average r between biological replicates (same haplotype) = 0.912, average r between haplotypes (i.e. between the wild-type and balancer) = 0.857. b-c, Zoomed-in versions of Fig. 3b, c. d, Location (promoter) and change in expression (balancer/wild-type, log2 fold change) of differentially expressed genes (DE-genes, orange, 23 genes), non-differentially expressed (non-DE, blue, 138 genes), lowly expressed (light-blue, 161), not testable due to lack of SNVs (grey, 121) genes within the 16 disrupted TADs. TADs (outlined in grey) are centered on the location of the inversion breakpoint, indicated by the vertical dashed line (purple). In the top six, the closest gene to the breakpoint is differentially expressed (orange), while the 10 TADs below the dashed line have one or more non-DE genes (blue, light blue) closer to the breakpoint. e, Expression levels of DE-genes and non-DE genes within disrupted TADs; FPKM, Fragments Per Kilobase of transcript per Million mapped reads.
Supplementary Figure 5 Characteristics of differential Hi-C and Capture-C contacts.
a-b, Top: positional distribution of differential Hi-C (a) or Capture-C (b) contacts originating from Transcription Start Sites (TSSs) of differentially expressed genes (DE-genes) with >1.5 fold change in expression (orange) and non-differentially expressed (non-DE, grey) genes. Genes are aligned by their TSS and transcribed to the right. 95% confidence bands (shaded ribbons) estimated using bootstrapping. Bottom right: heatmap of differential contacts (balancer/wild-type, log2 fold change) for 200 randomly sampled DE-genes (Hi-C) (a) or for all captured DE-genes (Capture C) (b). Bottom left: expression change (log2 fold change) for genes shown in the heatmap. c, Fraction of Hi-C (left) and Capture-C (right) differential contacts originating from different classes and having their other end overlapping a promoter of a differentially expressed or non-differentially expressed gene, a distal enhancer or a distal DHS. The observed fraction (purple) is compared to the distribution obtained from random shuffling (1,000 times) of differential contacts (grey). Center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range. p-values (two-sided permutation test) of significant differences (p<0.05) are indicated. Numbers in parentheses indicate: number of differential contacts in the given class, and (if applicable) number of unique genes, DE-genes or non-DE genes on the viewpoint side. d, Correlation of change in expression (log2 fold change) between pairs of DE-genes linked by a differential contact. Concordant gene expression (14 cases) in green, discordant (4 cases) in purple. Pearson correlation coefficient (r) indicated. e, Strength of all differential Capture-C contacts (balancer/wild-type, log2 fold change) established from each Capture-C probe region, ordered (left to right) based on the change in expression (log2 fold change) of the most DE gene assigned to the probe. Note that strongly differentially expressed genes (to the left and right of the purple arrows) do not have more, or stronger, differential Capture-C contacts.
Supplementary Figure 6 Nested inversions impact three neighboring loops.
a, Hi-C contact map (2 kb resolution, all Hi-C reads) showing long-range loops between Wbp2 and Nufip (top left), Wbp2 and kug (bottom left) and Nufip and kug (bottom right), indicated by asterisks. The locations of DNase hypersensitive sites (DHS), gene models, and differential gene expression (balancer/wild-type, log2 fold change) are shown. Differentially expressed genes (DE-genes) in orange, non-differentially expressed genes (non-DE) in blue and non-tested genes (lowly expressed or lacking SNVs) in grey. b-d, Zoomed-in wild-type (top) and balancer (bottom) Hi-C contact maps at 5 kb (left) and 20 kb (right) resolution showing the presence or absence of a long-range loop between Wbp2 and Nufip (b), Wbp2 and kug (c), Nufip and kug (d). e, Zoomed-out Hi-C contact map showing the presence or absence of long-range loops between these three loci in wild-type (bottom left) and balancer (top right) haplotypes. f, Schematic of the location and distance of Wbp2, Nufip and kug on chromosome 3 on wild-type (top) and balancer (bottom) haplotypes with respect to inversion breakpoints. Wbp2/Nufip are separated by similar distances in the wild-type (5.89 Mb) and balancer (5.34 Mb) chromosomes, and still form a loop despite intervening rearrangements (b). Wbp2/kug (c) and Nufip/kug (d) are separated by large distances in the balancer chromosome, leading to a severe diminishment of the looping interaction.
Supplementary information
Supplementary Information
Supplementary Figs. 1–6
Supplementary Table 1
PCR validation of randomly selected medium and large deletions
Supplementary Table 2
Exact balancer breakpoint coordinates
Supplementary Table 3
Hi-C read statistics
Supplementary Table 4
RNA-Seq library overview
Supplementary Table 5
Capture-C read statistics
Supplementary Table 6
Capture-C viewpoints
Supplementary Table 7
Differential Hi-C interactions between promoters of differentially expressed genes
Rights and permissions
About this article
Cite this article
Ghavi-Helm, Y., Jankowski, A., Meiers, S. et al. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat Genet 51, 1272–1282 (2019). https://doi.org/10.1038/s41588-019-0462-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-019-0462-3
This article is cited by
-
Transcriptionally active chromatin loops contain both ‘active’ and ‘inactive’ histone modifications that exhibit exclusivity at the level of nucleosome clusters
Epigenetics & Chromatin (2024)
-
Tissue-specific RNA Polymerase II promoter-proximal pause release and burst kinetics in a Drosophila embryonic patterning network
Genome Biology (2024)
-
Depletion of lamins B1 and B2 promotes chromatin mobility and induces differential gene expression by a mesoscale-motion-dependent mechanism
Genome Biology (2024)
-
Enhancer–promoter interactions become more instructive in the transition from cell-fate specification to tissue differentiation
Nature Genetics (2024)
-
Implications of the three-dimensional chromatin organization for genome evolution in a fungal plant pathogen
Nature Communications (2024)