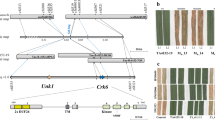
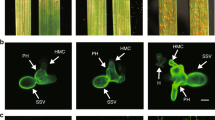
Abstract
Deployment of fast-evolving disease-resistance genes is one of the most successful strategies used by plants to fend off pathogens1,2. In gene-for-gene relationships, most cloned disease-resistance genes encode intracellular nucleotide-binding leucine-rich-repeat proteins (NLRs) recognizing pathogen-secreted isolate-specific avirulence (Avr) effectors delivered to the host cytoplasm3,4. This process often triggers a localized hypersensitive response, which halts further disease development5. Here we report the map-based cloning of the wheat Stb6 gene and demonstrate that it encodes a conserved wall-associated receptor kinase (WAK)-like protein, which detects the presence of a matching apoplastic effector6,7,8 and confers pathogen resistance without a hypersensitive response9. This report demonstrates gene-for-gene disease resistance controlled by this class of proteins in plants. Moreover, Stb6 is, to our knowledge, the first cloned gene specifying resistance to Zymoseptoria tritici, an important foliar fungal pathogen affecting wheat and causing economically damaging septoria tritici blotch (STB) disease10,11,12.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Michelmore, R. W., Christopoulou, M. & Caldwell, K. S. Impacts of resistance gene genetics, function, and evolution on a durable future. Annu. Rev. Phytopathol. 51, 291–319 (2013).
Krattinger, S. G. & Keller, B. Molecular genetics and evolution of disease resistance in cereals. New Phytol. 212, 320–332 (2016).
Jones, J. D. & Dangl, J. L. The plant immune system. Nature 444, 323–329 (2006).
Dodds, P. N. & Rathjen, J. P. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548 (2010).
Dickman, M. B. & Fluhr, R. Centrality of host cell death in plant-microbe interactions. Annu. Rev. Phytopathol. 51, 543–570 (2013).
Brading, P. A., Verstappen, E. C. P., Kema, G. H. J. & Brown, J. K. M. A gene-for gene relationship between wheat and Mycosphaerella graminicola, the Septoria tritici blotch pathogen. Phytopathology 92, 439–445 (2002).
Zhong, Z. et al. A small secreted protein in Zymoseptoria tritici is responsible for avirulence on wheat cultivars carrying the Stb6 resistance gene. New Phytol. 214, 619–631 (2017).
Kema, G.H.J. et al. Stress and sexual reproduction affect the dynamics of the wheat pathogen effector AvrStb6 and strobilurin resistance. Nat. Genet. https://doi.org/10.1038/s41588-018-0052-9 (2018).
Rudd, J. J., Keon, J. & Hammond-Kosack, K. E. The wheat mitogen-activated protein kinases TaMPK3 and TaMPK6 are differentially regulated at multiple levels during compatible disease interactions with Mycosphaerella graminicola. Plant Physiol. 147, 802–815 (2008).
Fones, H. & Gurr, S. The impact of Septoria tritici Blotch disease on wheat: An EU perspective. Fungal Genet. Biol. 79, 3–7 (2015).
Torriani, S. F. et al. Zymoseptoria tritici: a major threat to wheat production, integrated approaches to control. Fungal Genet. Biol. 79, 8–12 (2015).
Kettles, G. J. & Kanyuka, K. Dissecting the molecular interactions between wheat and the fungal pathogen Zymoseptoria tritici. Front. Plant Sci. 7, 508 (2016).
Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 35, 345–351 (2014).
Thomma, B. P., Nürnberger, T. & Joosten, M. H. Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23, 4–15 (2011).
Cook, D. E., Mesarich, C. H. & Thomma, B. P. H. J. Understanding plant immunity as a surveillance system to detect invasion. Annu. Rev. Phytopathol. 53, 541–563 (2015).
Sánchez-Vallet, A., McDonald, M. C., Solomon, P. S. & McDonald, B. A. Is Zymoseptoria tritici a hemibiotroph? Fungal Genet. Biol. 79, 29–32 (2015).
Kema, G. H. J., Yu, D. Z. & Rijkenberg, F. H. J. Histology of the pathogenesis of Mycosphaerella graminicola in wheat. Phytopathology 86, 777–786 (1996).
Torriani, S. F. F., Brunner, P. C., McDonald, B. A. & Sierotzki, H. QoI resistance emerged independently at least 4 times in European populations of Mycosphaerella graminicola. Pest Manag. Sci. 65, 155–162 (2009).
Cools, H. J. & Fraaije, B. A. Update on mechanisms of azole resistance in Mycosphaerella graminicola and implications for future control. Pest Manag. Sci. 69, 150–155 (2013).
Estep, L. K. et al. Emergence and early evolution of fungicide resistance in North American populations of Zymoseptoria tritici. Plant Pathol. 64, 961–971 (2015).
Brown, J. K., Chartrain, L., Lasserre-Zuber, P. & Saintenac, C. Genetics of resistance to Zymoseptoria tritici and applications to wheat breeding. Fungal Genet. Biol. 79, 33–41 (2015).
Tabib Ghaffary, S. M. et al. New broad-spectrum resistance to septoria tritici blotch derived from synthetic hexaploid wheat. Theor. Appl. Genet. 124, 125–142 (2012).
McDonald, B. A. & Mundt, C. C. How knowledge of pathogen population biology informs management of Septoria tritici blotch. Phytopathology 106, 948–955 (2016).
Chartrain, L., Brading, P. A. & Brown, J. K. M. Presence of the Stb6 gene for resistance to septoria tritici blotch (Mycosphaerella graminicola) in cultivars used in wheat-breeding programmes worldwide. Plant Pathol. 54, 134–143 (2005).
Arraiano, L. S. & Brown, J. K. M. Identification of isolate-specific and partial resistance to Septoria tritici blotch in 238 European wheat cultivars and breeding lines. Plant Pathol. 55, 726–738 (2006).
Arraiano, L. S. et al. Contributions of disease resistance and escape to the control of Septoria tritici blotch of wheat. Plant Pathol. 58, 910–922 (2009).
Ghaffary, S. M. et al. Genetic analysis of resistance to septoria tritici blotch in the French winter wheat cultivars Balance and Apache. Theor. Appl. Genet. 123, 741–754 (2011).
Saintenac, C., Jiang, D., Wang, S. & Akhunov, E. Sequence-based mapping of the polyploid wheat genome. G3 (Bethesda) 3, 1105–1114 (2013).
Bolot, S. et al. The ‘inner circle’ of the cereal genomes. Curr. Opin. Plant Biol. 12, 119–125 (2009).
Antolín-Llovera, M., Ried, M. K., Binder, A. & Parniske, M. Receptor kinase signaling pathways in plant-microbe interactions. Annu. Rev. Phytopathol. 50, 451–473 (2012).
Lee, W. S., Hammond-Kosack, K. E. & Kanyuka, K. Barley stripe mosaic virus-mediated tools for investigating gene function in cereal plants and their pathogens: virus-induced gene silencing, host-mediated gene silencing, and virus-mediated overexpression of heterologous protein. Plant Physiol. 160, 582–590 (2012).
Lee, W. S., Rudd, J. J. & Kanyuka, K. Virus induced gene silencing (VIGS) for functional analysis of wheat genes involved in Zymoseptoria tritici susceptibility and resistance. Fungal Genet. Biol. 79, 84–88 (2015).
McCallum, C. M., Comai, L., Greene, E. A. & Henikoff, S. Targeting induced local lesions in genomes (TILLING) for plant functional genomics. Plant Physiol. 123, 439–442 (2000).
King, R. et al. Mutation scanning in wheat by exon capture and next-generation sequencing. PLoS One 10, e0137549 (2015).
Krasileva, K. V. et al. Uncovering hidden variation in polyploid wheat. Proc. Natl. Acad. Sci. USA 114, E913–E921 (2017).
Dardick, C., Schwessinger, B. & Ronald, P. Non-arginine-aspartate (non-RD) kinases are associated with innate immune receptors that recognize conserved microbial signatures. Curr. Opin. Plant Biol. 15, 358–366 (2012).
Stotz, H. U., Mitrousia, G. K., de Wit, P. J. G. M. & Fitt, B. D. L. Effector-triggered defence against apoplastic fungal pathogens. Trends Plant Sci. 19, 491–500 (2014).
De Wit, P. J. G. M. Apoplastic fungal effectors in historic perspective; a personal view. New Phytol. 212, 805–813 (2016).
Diener, A. C. & Ausubel, F. M. RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 171, 305–321 (2005).
Hurni, S. et al. The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proc. Natl. Acad. Sci. USA 112, 8780–8785 (2015).
Zuo, W. et al. A maize wall-associated kinase confers quantitative resistance to head smut. Nat. Genet. 47, 151–157 (2015).
Brutus, A., Sicilia, F., Macone, A., Cervone, F. & De Lorenzo, G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. USA 107, 9452–9457 (2010).
Ferrari, S. et al. Oligogalacturonides: plant damage-associated molecular patterns and regulators of growth and development. Front. Plant Sci. 4, 49 (2013).
Kohorn, B. D. & Kohorn, S. L. The cell wall-associated kinases, WAKs, as pectin receptors. Front. Plant Sci. 3, 88 (2012).
Kohorn, B. D. Cell wall-associated kinases and pectin perception. J. Exp. Bot. 67, 489–494 (2016).
Park, A. R. et al. Interaction of the Arabidopsis receptor protein kinase Wak1 with a glycine-rich protein, AtGRP-3. J. Biol. Chem. 276, 26688–26693 (2001).
Shi, G. et al. The hijacking of a receptor kinase-driven pathway by a wheat fungal pathogen leads to disease. Sci. Adv. 2, e1600822 (2016).
Endo, T. R. & Gill, B. S. The deletion stocks of common wheat. J. Hered. 87, 295–307 (1996).
Sourdille, P. et al. An update of the Courtot x Chinese Spring intervarietal molecular marker linkage map for the QTL detection of agronomic traits in wheat. Theor. Appl. Genet. 106, 530–538 (2003).
Allen, A. M. et al. Transcript-specific, single-nucleotide polymorphism discovery and linkage analysis in hexaploid bread wheat (Triticum aestivum L.). Plant Biotechnol. J. 9, 1086–1099 (2011).
Wang, S. et al. Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol. J. 12, 787–796 (2014).
Keon, J. et al. Transcriptional adaptation of Mycosphaerella graminicola to programmed cell death (PCD) of its susceptible wheat host. Mol. Plant Microbe Interact. 20, 178–193 (2007).
Yuan, C. et al. A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS One 6, e26468 (2011).
Rieu, I. & Powers, S. J. Real-time quantitative RT-PCR: design, calculations, and statistics. Plant Cell 21, 1031–1033 (2009).
Lee, W.-S., Rudd, J. J., Hammond-Kosack, K. E. & Kanyuka, K. Mycosphaerella graminicola LysM effector-mediated stealth pathogenesis subverts recognition through both CERK1 and CEBiP homologues in wheat. Mol. Plant Microbe Interact. 27, 236–243 (2014).
Tassy, C., Partier, A., Beckert, M., Feuillet, C. & Barret, P. Biolistic transformation of wheat: increased production of plants with simple insertions and heritable transgene expression. Plant Cell Tissue Organ Cult. 119, 171–181 (2014).
Christensen, A. H. & Quail, P. H. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 5, 213–218 (1996).
Sparks, C. A. & Jones, H. D. Genetic transformation of wheat via particle bombardment. Methods Mol. Biol. 1099, 201–218 (2014).
Eriksen, L., Borum, F. & Jahoor, A. Inheritance and localisation of resistance to Mycosphaerella graminicola causing septoria tritici blotch and plant height in the wheat (Triticum aestivum L.) genome with DNA markers. Theor. Appl. Genet. 107, 515–527 (2003).
Chartrain, L., Brading, P. A., Widdowson, J. P. & Brown, J. K. Partial resistance to Septoria tritici blotch (Mycosphaerella graminicola) in wheat cultivars Arina and Riband. Phytopathology 94, 497–504 (2004).
Balfourier, F. et al. A worldwide bread wheat core collection arrayed in a 384-well plate. Theor. Appl. Genet. 114, 1265–1275 (2007).
Gouesnard, B. et al. MSTRAT: an algorithm for building germ plasm core collections by maximizing allelic or phenotypic richness. J. Hered. 92, 93–94 (2001).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011).
Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K. & Pease, L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 (1989).
Nallamsetty, S., Austin, B. P., Penrose, K. J. & Waugh, D. S. Gateway vectors for the production of combinatorially-tagged His6-MBP fusion proteins in the cytoplasm and periplasm of Escherichia coli. Protein Sci. 14, 2964–2971 (2005).
Taylor, I., Seitz, K., Bennewitz, S. & Walker, J. C. A simple in vitro method to measure autophosphorylation of protein kinases. Plant Methods 9, 22 (2013).
Kettles, G. J., Bayon, C., Canning, G., Rudd, J. J. & Kanyuka, K. Apoplastic recognition of multiple candidate effectors from the wheat pathogen Zymoseptoria tritici in the nonhost plant Nicotiana benthamiana. New Phytol. 213, 338–350 (2017).
Marín-de la Rosa, N. et al. Genome wide binding site analysis reveals transcriptional coactivation of cytokinin-responsive genes by DELLA proteins. PLoS Genet. 11, e1005337 (2015).
Acknowledgements
We thank the Biological Resource Centre on Small Grain Cereals (INRA, Clermont-Ferrand, France), the National Plant Germplasm System (USDA, US), the Germplasm Resources Unit (John Innes Centre, Norwich, UK), and the breeding companies KWS, Limagrain, RAGT Seeds, Saaten Union, Senova, Syngenta, Agri Obtentions, and Saatzucht Josef Breun GmbH & Co. KG for providing seed samples of different wheat species and varieties; E. Paux and H. Rimbert (INRA GDEC) for providing Stb6 expression data at different developmental stages; A. Doherty, A. Huttly, and C. Sparks (Rothamsted Research, Harpenden, UK) for vectors and wheat transformation; P. Isaac (iDNA Genetics Ltd., Norwich, UK) for transgene copy number analyses; INRA GDEC facilities for genotyping (GENTYANE) and wheat transformation; and S. Thomas (Rothamsted Research, Harpenden, UK) for providing vectors and protocols, and advice on the Y2H assay. We are also grateful to the International Wheat Genome Sequencing Consortium and K. Eversole for prepublication access to the IWGSC v1.0 wheat genome assembly. Research was funded by the Institute Strategic Program Grants ’20:20 Wheat’ (BB/J/00426×/1) and Designing Future Wheat (BB/P016855/1) from the Biotechnology and Biological Sciences Research Council of the UK (BBSRC) and the French National Institute for Agricultural Research (INRA).
Author information
Authors and Affiliations
Contributions
K.K., K.E.H.-K., C.S., and T.L. conceived the project. W.M. and H.B. screened the wheat BAC library. A.L.P., R.C.K., and C.U. provided the TILLING data. R.C.K. performed bioinformatics analyses and analyzed RNA-seq data. W.-S.L. performed VIGS. J.J.R. performed biochemical assays. S.J.P. performed statistical analysis. F.C., C.S., and K.K. carried out all other experiments and analyzed the data. K.K. and C.S. wrote the manuscript, and all authors revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
Rothamsted Research filed an International Patent Application (no. PCT/GB2016/053929 entitled ‘Plant Fungal Resistance Gene’) related to the content of this manuscript, on behalf of K.K., C.S., F.C., T.L., W.-S.L., and K.E.H.-K.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12, Supplementary Note and Supplementary Tables 1–10
Rights and permissions
About this article
Cite this article
Saintenac, C., Lee, WS., Cambon, F. et al. Wheat receptor-kinase-like protein Stb6 controls gene-for-gene resistance to fungal pathogen Zymoseptoria tritici. Nat Genet 50, 368–374 (2018). https://doi.org/10.1038/s41588-018-0051-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-018-0051-x
This article is cited by
-
The Egyptian wheat cultivar Gemmeiza-12 is a source of resistance against the fungus Zymoseptoria tritici
BMC Plant Biology (2024)
-
Transcriptomics of temperature-sensitive R gene-mediated resistance identifies a WAKL10 protein interaction network
Scientific Reports (2024)
-
Quantitative pathogenicity and host adaptation in a fungal plant pathogen revealed by whole-genome sequencing
Nature Communications (2024)
-
The ZmWAKL–ZmWIK–ZmBLK1–ZmRBOH4 module provides quantitative resistance to gray leaf spot in maize
Nature Genetics (2024)
-
Genome-wide identification and expression analysis of wall-associated kinase (WAK) and WAK-like kinase gene family in response to tomato yellow leaf curl virus infection in Nicotiana benthamiana
BMC Plant Biology (2023)