Abstract
We measured severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA concentrations in primary sewage sludge in the New Haven, Connecticut, USA, metropolitan area during the Coronavirus Disease 2019 (COVID-19) outbreak in Spring 2020. SARS-CoV-2 RNA was detected throughout the more than 10-week study and, when adjusted for time lags, tracked the rise and fall of cases seen in SARS-CoV-2 clinical test results and local COVID-19 hospital admissions. Relative to these indicators, SARS-CoV-2 RNA concentrations in sludge were 0–2 d ahead of SARS-CoV-2 positive test results by date of specimen collection, 0–2 d ahead of the percentage of positive tests by date of specimen collection, 1–4 d ahead of local hospital admissions and 6–8 d ahead of SARS-CoV-2 positive test results by reporting date. Our data show the utility of viral RNA monitoring in municipal wastewater for SARS-CoV-2 infection surveillance at a population-wide level. In communities facing a delay between specimen collection and the reporting of test results, immediate wastewater results can provide considerable advance notice of infection dynamics.
Similar content being viewed by others
Main
The progression of the COVID-19 pandemic has been monitored primarily by testing symptomatic individuals for the presence of SARS-CoV-2 RNA and counting the number of positive tests over time1. However, in the United States and other countries, the spread of COVID-19 has commonly exceeded the testing capacity of public health systems. Moreover, test results are a lagging indicator of the pandemic’s progression2,3, because testing is usually prompted by symptoms, which might take 2 weeks to present after infection4, and delays occur between the appearance of symptoms, testing and the reporting of test results. Monitoring sewage in a community’s collection or treatment system has been used previously to provide early surveillance of disease prevalence at a population-wide level, notably for polio5,6, and might be similarly beneficial for the current COVID-19 pandemic. SARS-CoV-2 RNA is present in the stool of patients with COVID-19 (refs. 7,8,9) and in raw wastewater10,11,12, and increased RNA concentrations in raw wastewater have been recently associated with increases in reported COVID-19 cases11. However, the utility of wastewater SARS-CoV-2 concentrations for tracking the progression of COVID-19 infections is poorly understood. In this study, we investigated how viral RNA concentrations in wastewater correlated with compiled testing and hospitalization data in a U.S. metropolitan area over ~10 weeks, corresponding to a first wave of SARS-CoV-2 infection.
When municipal raw wastewater discharges into treatment facilities, solids are settled and collected into a matrix called primary sewage sludge. We chose to analyze primary sludge rather than raw wastewater because it provides a high-solids-content, mixed sample that has been shown to contain a broad diversity of human viruses, including commonly circulating coronavirus strains13. During the COVID-19 outbreak, from March 19, 2020, to June 1, 2020, in the New Haven, Connecticut, metropolitan area, we collected daily primary sludge samples from the wastewater treatment facility, which serves ~200,000 residents. We quantitatively compared SARS-CoV-2 RNA concentrations in sludge with publicly reported data on four other measures of the outbreak: SARS-CoV-2 positive test results by date of specimen collection; the percentage of positive SARS-CoV-2 test results (test positivity) by date of specimen collection; the number of local hospital admissions of patients with COVID-19; and SARS-CoV-2 positive test results by reporting date.
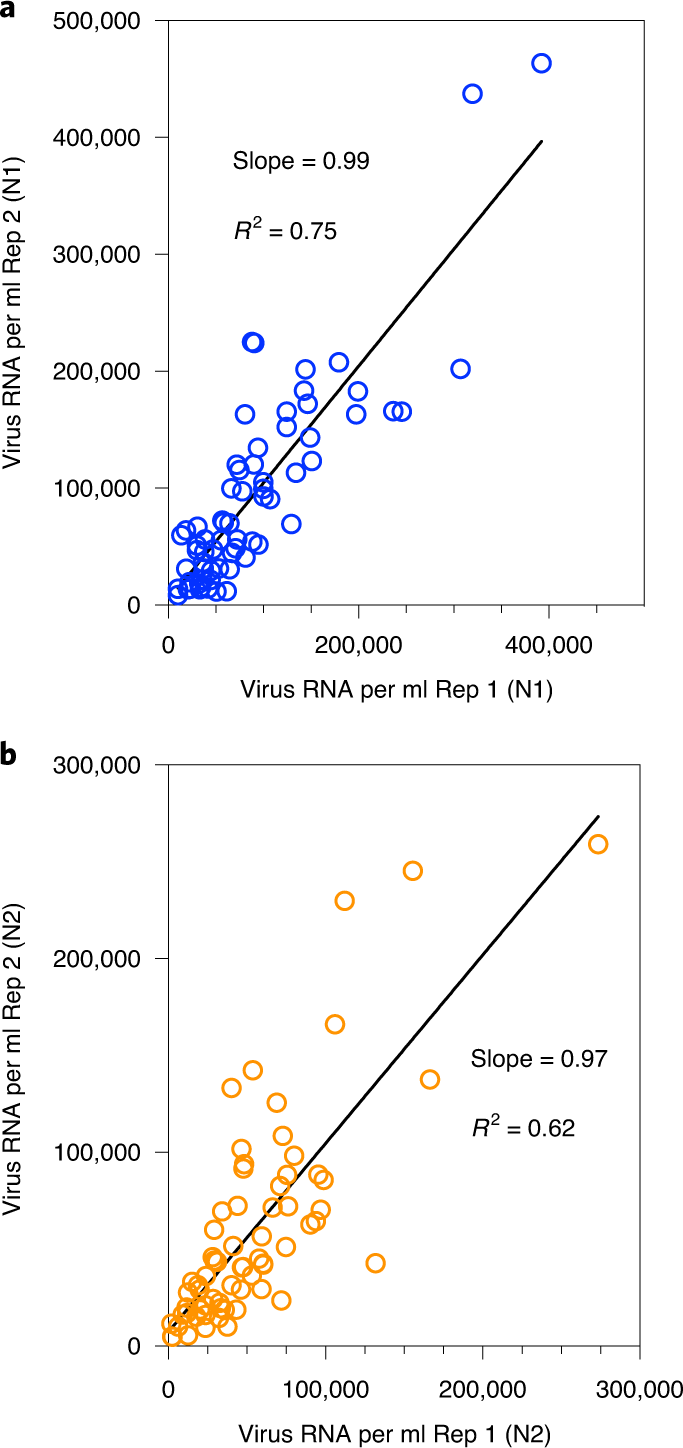
We measured SARS-CoV-2 virus RNA by quantitative reverse transcription polymerase chain reaction (qRT–PCR) using the same N1 and N2 primer sets employed in COVID-19 individual testing. Virus RNA copies ranged from 1.7 × 103 ml−1 to 4.6 × 105 ml−1 of primary sludge. All qRT–PCR concentration threshold (Ct) values were below 40, and 97% of all samples had a Ct value less than 38. The average Ct was 34.6 for N1 primers and 34.5 for N2 primers. Values for each replicate were reported as positive only when the human ribonuclease P (RP) internal control gene was positive. The average (s.d.) Ct value for the RP gene for all positive samples was 36.2 (1.2) for replicate 1 and 36.2 (1.3) for replicate 2. Replicated samples demonstrated similar SARS-CoV-2 RNA concentration values (Fig. 1). Concentration comparisons between replicates produced slopes of 0.99 (R2 = 0.75) for N1 primers and 0.97 (R2 = 0.62) for N2 primers.
All five measures traced the rise and fall of SARS-CoV-2 infections during the more than 10-week period studied (Fig. 2). However, the sludge results showed an increase during the first week (March 19–25, 2020) that was not observed in the reported testing or hospital admissions data. Applying a distributed lag measurement error time series model allowed an estimation of relationships between viral time series results and the reported testing and hospital admissions data. By modeling the epidemiological time series as a function of the sludge SARS-CoV-2 RNA data across multiple daily lags (posterior means ± 90% credible intervals), we found that the sludge results led the number of positive tests by date of specimen collection by 0–2 d, with a potential lag of 1 d (Fig. 3a,b); the percentage of positive tests by date of specimen collection by 0–2 d, with a potential lag of 1 d (Fig. 3c,d); hospital admissions by 1–4 d (Fig. 3e,f); and the number of positive tests by report date by 6–8 d (Fig. 3g,h). Performing the time series analysis with or without adjustment for testing volume did not result in differences in estimated lag times between sludge viral RNA results and number of positive tests (based on the above date of specimen collection results and date reported to the Connecticut State Department of Public Health) (Extended Data Fig. 1).
All data represent the cities of New Haven, Hamden, East Haven and Woodbridge, Connecticut, which are served by the ESWPAF. The blue vertical dashed lines indicate the first week of analysis, March 19–25, 2020. a,b, Number of positive SARS-CoV-2 test results, reported by date of specimen collection. c,d, Percentage of positive SARS-CoV-2 test results, reported by date of specimen collection. e,f, Number of COVID-19 admissions to Yale New Haven Hospital for residents of the four cities. g,h, Number of positive SARS-CoV-2 test results by public reporting date. i,j, Primary sludge SARS-CoV-2 RNA concentration (virus RNA gene copies per ml of sludge).
a, Daily lags of 0–2 d and leads of 1 d are associated with the number of positive tests based on specimen collection date. b, Cumulative relationship between viral RNA in sludge and the number of positive tests based on specimen collection date. c, Daily lags of 0–2 d and leads of 1 d are associated with the percentage of positive tests based on specimen collection date. d, Cumulative relationship between viral RNA in sludge and the percentage of positive tests based on specimen collection date. e, Daily lags of 1–4 d are associated with hospitalization. f, Cumulative relationship between viral RNA in sludge and hospital admissions. g, Daily lags of sludge virus RNA data at longer time lags (6–8 d in the past) best correlate with the time series of publicly reported positive tests. h, Cumulative beta relationship between viral RNA in sludge and reported number of positive tests. Posterior means at the center of each data point and 90% credible intervals for error bars are displayed. For each lag, n = 75 daily values for positive tests by date of specimen collection (a,b), 75 daily values for percentage of positive tests by date of specimen collection (c,d), 75 daily values for hospital admission (e,f) and 75 daily values for publicly reported positive tests (g,h).
Overall, our results demonstrate that measurement of SARS-CoV-2 RNA concentrations in primary sludge provides an approach to estimate changes in COVID-19 prevalence on a population level. Sludge results were not a leading indicator compared to positive test results or percentage of positive tests by date of specimen collection. However, they led hospitalizations by 1–4 d and test results by report date by ~1 week. Thus, in communities where test reporting is delayed, sludge results, if analyzed and reported on the same day as sampling, can provide substantial advance notice of infection dynamics. In locations with rapid reporting of SARS-CoV-2 test results, the lead time afforded by sewage surveillance might be significantly reduced. The lags in test reporting have multiple causes and might vary with societal responses to the pandemic. COVID-19 arrived in the New Haven metropolitan area in early March 2020, when testing capacity was limited, and there were extended waiting times from test date to reporting date. Understanding and mitigating the causes of such lags will require additional research.
Sludge data are also susceptible to variability for multiple reasons. For example, primary sludge handling approaches are specific to particular treatment plants and could affect the levels of detectable virus. Given the uncertainties in sludge data and epidemiological data, we did not attempt to correlate absolute numbers of sludge SARS-CoV-2 RNA concentrations and COVID-19 cases.
Monitoring primary sludge is a broadly applicable strategy. Wastewater treatment plants with primary and secondary treatment are standard in many regions of the world, and treatment facilities are rapidly expanding in urban areas of lower- and middle-income countries14. In the United States, approximately 16,000 treatment plants serve more than 250,000,000 people. In regions without primary wastewater treatment, monitoring of raw wastewater streams would be necessary. Our results indicate that jurisdictions can use primary sludge SARS-CoV-2 concentrations as an additional basis for imposing or easing infection-control restrictions, especially in locations affected by limits in clinical testing capacity or delays in test reporting.
Methods
Sample collection
Primary sewage sludge (40 ml) was collected from the East Shore Water Pollution Abatement Facility (ESWPAF) in New Haven, Connecticut, USA. A total of 73 samples were taken daily from March 19, 2020, to June 1, 2020, between 8:00 and 10:00 EDT, and stored at −80 °C before analysis (samples were not available on May 3 and 6). The first sampling dates were before widespread testing in the region and before the March 23, 2020, stay-at-home restrictions implemented throughout the State of Connecticut. From the sampling start and end dates, cities served by the ESWPAF experienced an increase in confirmed COVID-19 cases (by testing) from seven cases to 3,978 (ref. 15). The plant serves an estimated population of 200,000 people with average treated flows of 1.75 m3 s−1. Sludge collected from ESWPAF is primary sludge, sampled at the outlet of a gravity thickener, ranging in solids content from 2.6% to 5%. The solids residence time in the gravity thickener is 4 h.
Viral RNA quantitative testing
To quantify SARS-CoV-2 RNA concentrations in primary sludge, 2.5 ml of well-mixed sludge samples were added directly to a commercial kit optimized for isolation of total RNA from soil (RNeasey PowerSoil Total RNA Kit, Qiagen). Two replicate RNA extractions and analyses were performed for one daily primary sludge sample. Isolated RNA pellets were dissolved in 50 μl of ribonuclease-free water, and total RNA was measured by spectrophotometry (NanoDrop, Thermo Fisher Scientific). SARS-CoV-2 RNA was quantified by one-step qRT–PCR using the U.S. Centers for Disease Control and Prevention (CDC) N1 and N2 primers sets16,17. For control and in accordance with the CDC protocol, analysis was also conducted for the human RP gene17, and SARS-CoV-2 results were reported only if RP detection was positive. Samples were analyzed using the Bio-Rad iTaq Universal Probes One-Step Kit in 20-µl reactions run at 50 °C for 10 min and 95 °C for 1 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s per the manufacturer’s recommendations. SARS-CoV-2 RNA concentrations were determined using a standard curve as previously described17 and presented as virus RNA copies. For the standard curve, complementary DNA synthesized from full-length SARS-CoV-2 RNA (WA1-USA strain) was used as a template to generate SARS CoV-2 N gene transcripts as previously described17. The N gene was amplified, and the PCR amplicon was purified and used for template in the MEGAscript T7 Kit (Thermo Fisher Scientific) to generate single-stranded RNA transcripts. RNA was quantified on a Qubit fluorimeter (Thermo Fisher Scientific) and integrity verified on a Bioanalyzer 2100 (Agilent). Viral RNA copies were calculated, and serial ten-fold dilutions were made. To validate our N1 and N2 primers sets, standard curves using the ten-fold series dilution (5 × 101 to 5 × 108 copies per reaction) of the N gene transcripts were analyzed. The N1 primer set generated a standard curve with an R2 value of 0.98 with an efficiency of 94.1% (slope = −3.473; y intercept = 42.266). The N2 primer set generated a standard curve with an R2 value of 0.99 with an efficiency of 88.5% (slope = −3.632; y intercept = 42.528).
The SARS-CoV-2 concentration results were adjusted to the total RNA extracted by multiplying sample concentrations by the ratio of the maximum RNA concentration to the sample RNA concentration. This accounts for day-to-day variations in sludge solids content and RNA extraction efficiency. To determine whether sludge RNA extracts contained PCR inhibition, target RNA was spiked into three separate sterile, ribonuclease-free water samples (no inhibition) and five different sludge RNA extracts from samples collected at a time in the outbreak when cases were low and viral RNA was not detected with N1 primers. Spiked samples were then diluted 5× and 25×, and sludge RNA Ct values were compared to water RNA Ct values using N1 primers. No differences were observed for average water Ct values and sludge extract Ct vales for the no dilution (P = 0.14), 5× dilution (P = 0.51) and 25× dilution (P = 0.23), two-tailed t-test, suggesting no PCR inhibition in the RNA extracts. All samples were diluted 5× for use as a template to ensure that qRT–PCR inhibition occurred. Sewage sludge from March 2018 was used as a control, and no SARS-CoV-2 detection was observed from either N1 or N2 primers. These control sludges were stored at −80 °C and were consistently positive for the human RP gene. Positive RNA controls and no-template controls were included in all qRT–PCR runs. Appropriate Ct values were observed for all positive controls, and no amplification was observed in negative template controls.
Epidemiological data
Daily COVID-19 admissions to the Yale New Haven Hospital were compiled from hospital records—adjusted to include only New Haven, East Haven, Hamden and Woodbridge, Connecticut residents—and confirmed by laboratory testing. Hospital data were obtained from the Joint Data Analytics Team for the New Haven Health System. The total number and percentage of tests of residents from the four cities that were positive for COVID-19 and reported by date of specimen collection and date of reporting to the State of Connecticut were supplied through a data request to the Connecticut Department of Public Health. Numbers of laboratory-confirmed positive COVID-19 tests by report date in the towns served by the ESWPAF (New Haven, East Haven, Hamden and Woodbridge, Connecticut) were compiled from daily reports published by the Connecticut Department of Public Health15.
Statistics
Linear regressions were used to estimate the relationship between SARS-CoV-2 RNA copies per ml results for replicated RNA extractions of each daily sample (n = 73 for each PCR primer). Two-tailed t-tests (α = 0.05) were used in PCR inhibition experiments to determine if spiked sludge RNA extracts resulted in the same Ct values as spiked water samples at no dilution, 5× dilution and 25× dilution, n = 6 for water spiked samples and n = 15 for sludge spiked samples for each dilution.
Estimation of primary sludge as a potential leading indicator was performed using a distributed lag measurement error time series model. This analysis was carried out in the Bayesian framework, allowing us to correctly characterize multiple sources of uncertainty when estimating the lagged associations of interest. In the analyses, we assume that the observed sludge testing data represent unbiased estimates of an underlying, unobserved trajectory of viral concentration in the sludge. We then evaluated the association between the underlying trajectory of viral concentration in the sludge at multiple lagged periods and the number of positive tests (based on date of specimen collection)/percentage of positive tests based on date of specimen collection)/hospitalizations/number of positive tests (based on date of report) using distributed lag Poisson regression models that included a random effect to account for overdispersion and autocorrelation in the outcome, n = 73 for sludge virus RNA samples, n = 75 daily positive tests by date of specimen collection, n = 75 daily percentage of positive tests reported by date of specimen collection, n = 75 daily reported hospitalizations and n = 75 daily reported positive tests. The distributed lag regression parameters were modeled using a random walk process. The models are fit using the rjags package in R18. Mathematical models include the following:
Model for case counts:
Model for primary sludge:
Prior distributions:
where d is the number of past lags included in the model, n is the total number of days of available data, m is the number of primary sludge replicates on a given day, Yt is the case count on day t, Ot is the offset on day t (number of tests performed on each day for the analysis of cases by test date or 0 otherwise), Wtj is the measured concentration of virus in sludge from sample replicate j on day t, and xt is the unobserved true concentration of virus in sludge on day t.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The epidemiological data used in this study are provided as Source Data to Fig. 2. All sludge data are available upon request.
References
Dong, E., Du, H. & Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 20, 533–534 (2020).
Mizumoto, K., Kagaya, K., Zarebski, A. & Chowell, G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Euro. Surveill. 25, 2000180 (2020).
García-Basteiro, A. L. et al. Monitoring the COVID-19 epidemic in the context of widespread local transmission. Lancet Respir. Med. 8, 440–442 (2020).
Lauer, S. A. et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Intern. Med. 172, 577–582 (2020).
Pöyry, T., Stenvik, M. & Hovi, T. Viruses in sewage waters during and after a poliomyelitis outbreak and subsequent nationwide oral poliovirus vaccination campaign in Finland. Appl. Environ. Microbiol. 54, 371–374 (1988).
Berchenko, Y. et al. Estimation of polio infection prevalence from environmental surveillance data. Sci. Transl. Med. 9, eaaf6786 (2017).
Wang, W. et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 323, 1843–1844 (2020).
Xu, Y. et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 26, 502–505 (2020).
Zhang, J., Wang, S. & Xue, Y. Fecal specimen diagnosis 2019 novel coronavirus-infected pneumonia. J. Med. Virol. 92, 680–682 (2020).
Ahmed, W. et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 728, 138764 (2020).
Medema, G., Heijnen, L., Elsinga, G., Italiaander, R. & Brouwer, A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 7, 511–516 (2020).
Wu, F. et al. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems 5, e00614–e00620 (2020).
Bibby, K. & Peccia, J. Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environ. Sci. Technol. 47, 1945–1951 (2013).
Zhang, Q. H. et al. Current status of urban wastewater treatment plants in China. Environ. Int. 92-93, 11–22 (2016).
State of Connecticut, CT Data: Confirmed COVID-19 Cases. Connecticut Department of Health https://data.ct.gov/Health-and-Human-Services/COVID-19-confirmed-cases-by-town-/28fr-iqnx (2020).
U.S. Centers for Disease Control and Prevention. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT–PCR Diagnostic Panel https://www.fda.gov/media/134922/download?fbclid=IwAR1DdEweazD3ixmrpZMc07VXM0_n1qx455rGV7E0fAEcA1QZf3Peh0Qxypo (2020).
Vogels, C. B. F. et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat. Microbiol. https://doi.org/10.1038/s41564-020-0761-6 (2020).
Plummer, M. JAGS: a program for analysis of Bayesian graphical models using gibbs sampling. In Proc. 3rd International Workshop on Distributed Statistical Comptuting (DSC, 2003).
Acknowledgements
We wish to thank the Greater New Haven Water Pollution Control Authority, New Haven, Connecticut, USA, for primary sludge sampling assistance. A.Z. is supported by a CoReCT seed grant from the Yale School of Medicine and the Yale Institute of Global Health. N.D.G. is supported by a gift from the Huffman Family Donor Advised Fund. A.K. and A.C.-M. are supported by the Beatrice Kleinberg Neuwirth Fund, the Yale School of Public Health and the National Institutes of Health (R01 AI121207). The authors thank K. Yousey-Hindes and P. Clogher for providing the data on percentage of positive tests.
Author information
Authors and Affiliations
Contributions
J.P., A.Z., D.E.B., N.D.G., D.W., M.W. and W.A. contributed laboratory analysis and sample collection. J.P., E.H.K., A.C.-M., A.I.K., A.A.M., J.L.W., D.M.W. and S.B.O. contributed epidemiological analysis. D.M.W. and J.L.W. produced the distributed lag analysis.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Estimated daily distributed lag parameters describing the association between viral RNA in sludge and positive COVID-19 tests, based on date of report to CT Department of Health.
The top row figures are adjusted for testing volume, bottom row figures are unadjusted for testing volume. a, daily lags of sludge data at longer time lags (0 to 4 days in the past) best correlate with the time series of reported positive tests (adjusted). b, cumulative relationship between viral RNA in sludge and number of positive tests (adjusted). c, daily lags of sludge data at longer time lags (0 to 4 days in the past) best correlate with the time series of reported cases (not adjusted for number of tests). d, cumulative relationship between viral RNA in sludge and number of positive tests (not adjusted). The posterior means at the center of each data point and 90% credible intervals for error bars are displayed. For each lag, n=73 daily virus RNA samples, n=75 daily reported adjusted positive test values, n=75 daily reported unadjusted positive test values.
Supplemantary information
Source data
Source Data Fig. 2
Number of positive COVID-19 tests by date of specimen collection, percentage of positive COVID-19 tests by date of specimen collection, hospital admissions for patients with COVID-19 and number of COVID-19 positive tests by date of report. All data are for New Haven, Hamden, East Haven and Woodbridge, Connecticut, USA.
Rights and permissions
About this article
Cite this article
Peccia, J., Zulli, A., Brackney, D.E. et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat Biotechnol 38, 1164–1167 (2020). https://doi.org/10.1038/s41587-020-0684-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-020-0684-z
This article is cited by
-
A broad wastewater screening and clinical data surveillance for virus-related diseases in the metropolitan Detroit area in Michigan
Human Genomics (2024)
-
Emergence and spread of SARS-CoV-2 variants of concern in Canada: a retrospective analysis from clinical and wastewater data
BMC Infectious Diseases (2024)
-
New prospects of environmental RNA metabarcoding research in biological diversity, ecotoxicological monitoring, and detection of COVID-19: a critical review
Environmental Science and Pollution Research (2024)
-
Low-cost and rapid sensors for wastewater surveillance at low-resource settings
Nature Water (2023)
-
Monitoring of over-the-counter (OTC) and COVID-19 treatment drugs complement wastewater surveillance of SARS-CoV-2
Journal of Exposure Science & Environmental Epidemiology (2023)