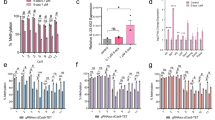
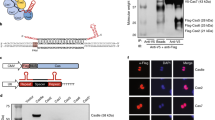
Abstract
Gene expression can be activated or suppressed using CRISPR–Cas9 systems. However, tools that enable dose-dependent activation of gene expression without the use of exogenous transcription regulatory proteins are lacking. Here we describe chemical epigenetic modifiers (CEMs) designed to activate the expression of target genes by recruiting components of the endogenous chromatin-activating machinery, eliminating the need for exogenous transcriptional activators. The system has two parts: catalytically inactive Cas9 (dCas9) in complex with FK506-binding protein (FKBP) and a CEM consisting of FK506 linked to a molecule that interacts with cellular epigenetic machinery. We show that CEMs upregulate gene expression at target endogenous loci up to 20-fold or more depending on the gene. We also demonstrate dose-dependent control of transcriptional activation, function across multiple diverse genes, reversibility of CEM activity and specificity of our best-in-class CEM across the genome.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Genome-wide data generated herein are publicly available through the Gene Expression Omnibus with accession number GSE129407. All data presented in this manuscript are available from the corresponding authors upon reasonable request.
Code availability
Readers can view our code through the public link (https://github.com/GryderArt/ChIPseqPipe). There are no access restrictions.
References
Dawson, M. A. The cancer epigenome: concepts, challenges, and therapeutic opportunities. Science 355, 1147–1152 (2017).
MacDonald, I. A. & Hathaway, N. A. Epigenetic roots of immunologic disease and new methods for examining chromatin regulatory pathways. Immunol. Cell Biol. 93, 261–270 (2015).
Zeng, L. & Zhou, M. M. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 513, 124–128 (2002).
de Ruijter, A. J. M., van Gennip, A. H., Caron, H. N., Kemp, S. & van Kuilenburg, A. B. P. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370, 737–749 (2003).
Gao, Y. et al. Complex transcriptional modulation with orthogonal and inducible dCas9 regulators. Nat. Methods 13, 1043–1049 (2016).
Chen, T. et al. Chemically controlled epigenome editing through an inducible dCas9 system. J. Am. Chem. Soc. 139, 11337–11340 (2017).
Braun, S. M. G. et al. Rapid and reversible epigenome editing by endogenous chromatin regulators. Nat. Commun. 8, 560 (2017).
Gao, D. & Liang, F. S. Chemical inducible dCas9-guided editing of H3K27 acetylation in mammalian cells. Methods Mol. Biol. 1767, 429–445 (2018).
Ma, D., Peng, S. & Xie, Z. Integration and exchange of split dCas9 domains for transcriptional controls in mammalian cells. Nat. Commun. 7, 13056 (2016).
Shrimp, J. H. et al. Chemical control of a CRISPR–Cas9 acetyltransferase. ACS Chem. Biol. 13, 455–460 (2018).
Hilton, I. B. et al. Epigenome editing by a CRISPR–Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 33, 510–517 (2015).
Liszczak, G. P. et al. Genomic targeting of epigenetic probes using a chemically tailored Cas9 system. Proc. Natl Acad. Sci. USA 114, 681–686 (2017).
Erwin, G. S. et al. Synthetic transcription elongation factors license transcription across repressive chromatin. Science 6370, 1617–1622 (2017).
Butler, K. V., Chiarella, A. M., Jin, J. & Hathaway, N. A. Targeted gene repression using novel bifunctional molecules to harness endogenous histone deacetylation activity. ACS Synth. Biol. 7, 38–45 (2018).
Chung, C. et al. Discovery and characterization of small molecule inhibitors of the BET family bromodomains. J. Med. Chem. 54, 3827–3838 (2011).
Demont, E. H. et al. 1,3-Dimethyl benzimidazolones are potent, selective inhibitors of the BRPF1 bromodomain. ACS Med. Chem. Lett. 5, 1190–1195 (2014).
Hay, D. A. et al. Discovery and optimization of small-molecule ligands for the CBP/p300 bromodomain. J. Am. Chem. Soc. 26, 9308–9319 (2014).
Chavez, A. et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12, 326–328 (2015).
Morita, S. et al. Targeted DNA demethylation in vivo using dCas9–peptide repeat and scFv–TET1 catalytic domain fusions. Nat. Biotechnol. 34, 1060–1065 (2016).
Tanenbaum, M. E., Gilbert, L. A., Qi, L. S., Weissman, J. S. & Vale, R. D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646 (2014).
Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR–Cas9 complex. Nature 517, 583–588 (2015).
Lim, F. et al. Altering the RNA binding specificity of a translational repressor. J. Biol. Chem. 12, 9006–9010 (1994).
Chavez, A. et al. Comparison of Cas9 activators in multiple species. Nat. Methods 13, 563–567 (2016).
Begley, C. G. & Ellis, L. M. Raise standards for preclinical cancer research. Nature 483, 531–533 (2012).
Hay, M., Thomas, D. W., Craighead, J. L., Economides, C. & Rosenthal, J. Clinical development success rates for investigational drugs. Nat. Biotechnol. 32, 40–51 (2014).
Gryder, B. E. et al. PAX3–FOXO1 establishes myogenic super enhancers and confers BET bromodomain vulnerability. Cancer Discov. 7, 884–899 (2017).
Lovén, J. et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013).
Lin, X. et al. HEXIM1 as a robust pharmacodynamic marker for monitoring target engagement of BET family bromodomain inhibitors in tumors and surrogate tissues. Mol. Cancer Ther. 16, 388–396 (2017).
Hathaway, N. A. et al. Dynamics and memory of heterochromatin in living cells. Cell 149, 1447–1460 (2012).
Thakore, P. I. et al. RNA-guided transcriptional silencing in vivo with S. aureus CRISPR–Cas9 repressors. Nat. Commun. 9, 1674 (2018).
Liu, S. et al. Editing DNA methylation in the mammalian genome. Cell 167, 233–247 (2016).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Chiarella, A. M. et al. Cavitation enhancement increases the efficiency and consistency of chromatin fragmentation from fixed cells for downstream quantitative applications. Biochemistry 57, 2756–2761 (2018).
Acknowledgements
We thank the members of the Hathaway, Jin and Khan laboratories for many helpful discussions. We acknowledge E. Chory (Stanford University), S. Braun (Stanford University), G. Crabtree (Stanford University), R. Vale (University of California, San Francisco), F. Zhang (Massachusetts Institute of Technology), Z. Xie (Tsinghua University) and C. Gersbach (Duke University) for sharing plasmids used or adapted in this study. This work was supported in part by grant R01GM118653 from the US National Institutes of Health (to N.A.H.) and by grants R01GM122749 and R01HD088626 from the US National Institutes of Health (to J.J.). This work was further supported by a Tier 3 grant and a Student Grant from the UNC Eshelman Institute for Innovation (to N.A.H. and A.M.C., respectively). T32GM007092 from the US National Institutes of Health (to A.M.C.) also supported early portions of this work. Additional support was provided by American Cancer Society postdoctoral fellowship PF-14-021-01-CDD (to K.V.B.). Flow cytometry data were obtained at the UNC Flow Cytometry Core Facility, funded by the P30CA016086 Cancer Center Core Support Grant to the UNC Lineberger Comprehensive Cancer Center.
Author information
Authors and Affiliations
Contributions
N.A.H., J.J., K.V.B. and A.M.C. conceived the project. N.A.H., J.J., K.V.B., A.M.C., D.L., B.E.G. and J.K. contributed to the experimental design. N.A.H., J.J., K.V.B., A.M.C., D.L., B.E.G., J.K., T.A.W., X.Y. and S.P. contributed experimentally. K.V.B. synthesized the compounds. B.E.G. and J.K. carried out ChIP–seq and RNA-seq experiments and analysis. N.A.H., J.J., K.V.B., A.M.C., D.L. and B.E.G. wrote the manuscript. All authors edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
N.A.H., J.J., K.V.B. and A.M.C. are inventors on U.S. provisional patent application no. 62/654,958, “Bifunctional chemical epigenetic modifiers and methods of use.”
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Materials
Supplementary Figures 1–5, Supplementary Tables 1–3 and Supplementary Note 1.
Rights and permissions
About this article
Cite this article
Chiarella, A.M., Butler, K.V., Gryder, B.E. et al. Dose-dependent activation of gene expression is achieved using CRISPR and small molecules that recruit endogenous chromatin machinery. Nat Biotechnol 38, 50–55 (2020). https://doi.org/10.1038/s41587-019-0296-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-019-0296-7
This article is cited by
-
Context-specific functions of chromatin remodellers in development and disease
Nature Reviews Genetics (2024)
-
CasTuner is a degron and CRISPR/Cas-based toolkit for analog tuning of endogenous gene expression
Nature Communications (2023)
-
Precise programming of multigene expression stoichiometry in mammalian cells by a modular and programmable transcriptional system
Nature Communications (2023)
-
Histone post-translational modifications — cause and consequence of genome function
Nature Reviews Genetics (2022)
-
CRISPR technologies for precise epigenome editing
Nature Cell Biology (2021)