Abstract
Single-cell RNA sequencing can reveal the transcriptional state of cells, yet provides little insight into the upstream regulatory landscape associated with open or accessible chromatin regions. Joint profiling of accessible chromatin and RNA within the same cells would permit direct matching of transcriptional regulation to its outputs. Here, we describe droplet-based single-nucleus chromatin accessibility and mRNA expression sequencing (SNARE-seq), a method that can link a cell’s transcriptome with its accessible chromatin for sequencing at scale. Specifically, accessible sites are captured by Tn5 transposase in permeabilized nuclei to permit, within many droplets in parallel, DNA barcode tagging together with the mRNA molecules from the same cells. To demonstrate the utility of SNARE-seq, we generated joint profiles of 5,081 and 10,309 cells from neonatal and adult mouse cerebral cortices, respectively. We reconstructed the transcriptome and epigenetic landscapes of major and rare cell types, uncovered lineage-specific accessible sites, especially for low-abundance cells, and connected the dynamics of promoter accessibility with transcription level during neurogenesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Raw and processed data are available at the Gene Expression Omnibus database under accession number GSE126074.
Code availability
The custom script for processing single-nucleus chromatin accessibility reads is available at https://github.com/chensong611/SNARE_prep.
References
Lake, B. B. et al. Nat. Biotechnol. 36, 70–80 (2018).
Duren, Z. et al. Proc. Natl Acad. Sci. USA 115, 7723–7728 (2018).
Macosko, E. Z. et al. Cell 161, 1202–1214 (2015).
Amini, S. et al. Nat. Genet. 46, 1343–1349 (2014).
Cao, J. et al. Science 361, 1380–1385 (2018).
Buenrostro, J. D. et al. Nat. Methods 10, 213–218 (2013).
Corces, M. R. et al. Nat. Methods 14, 959–962 (2017).
Zamanighomi, M. et al. Nat. Commun. 9, 2410 (2018).
Rosenberg, A. B. et al. Science 360, 176–182 (2018).
Preissl, S. et al. Nat. Neurosci. 21, 432–439 (2018).
Yuzwa, S. A. et al. Cell Rep. 21, 3970–3986 (2017).
Bravo González-Blas, C. et al. Nat. Methods 16, 397–400 (2019).
Subramanian, L. et al. Proc. Natl Acad. Sci. USA 108, E265–E274 (2011).
Zhang, S. et al. Mol. Neurobiol. 55, 9001–9015 (2018).
Harrison-Uy, S. J. & Pleasure, S. J. Cold Spring Harb. Perspect. Biol. 4, a008094 (2012).
Artegiani, B. et al. Cell. Rep. 21, 3271–3284 (2017).
La Manno, G. et al. Nature 560, 494–498 (2018).
Lee, K. E. et al. Proc. Natl Acad. Sci. USA 111, 2794–2799 (2014).
Iijima, T. et al. Cell 147, 1601–1614 (2011).
Acknowledgements
This project was supported by National Institutes of Health grants U01MH098977, R01HL123755 and U54HL145608 to K.Z.
Author information
Authors and Affiliations
Contributions
S.C. and K.Z. conceived the study. S.C. designed and conducted the experiments. S.C. and B.B.L. analyzed the data. S.C., B.B.L. and K.Z. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Quality metrics of SNARE-seq chromatin profiles of GM12878 cells.
a, DNA fragment size distribution showing nucleosomal periodicity. (Inset) log-transformed histogram. b, Representative Venn diagram showing the numbers of overlapping accessible sites generated using ATAC-seq, Omni-ATAC and SNARE-seq chromatin assays. c, Enrichment of chromatin accessibility signals around transcription start sites (TSS). Top: enrichment around individual TSS. Bottom: aggregated enrichment across all TSSs. X axis indicates the relative distance to TSSs. d, Total number of accessible sites versus fraction of reads in open chromatin peaks (FRiP) within GM12878 cells (n = 3,787 after filtration). Dotted lines (2,000 and 49%) represent cutoffs used for downstream analysis.
Supplementary Figure 2 Comparison of the number of accessible sites, genes and transcripts detected per nucleus by SNARE-seq with different single-cell/nucleus chromatin accessibility or RNA-seq methods.
a, Histogram showing the numbers of accessible sites captured by SNARE-seq chromatin profiles. b, Histogram showing the numbers of accessible sites detected per nucleus with different single-cell/nucleus ATAC-seq methods. The processed peak count matrices of published reports were downloaded from GEO (sc-ATAC, GSE65360; sci-ATAC, GSE68103; snATAC, GSE100033; sci-CAR, GSE117089) and binarized. c, Histogram showing the fraction of reads in peaks (FRiP) within GM12878 or postnatal day 0 mouse cerebral cortex SNARE-seq chromatin accessibility data. GM12878, GM; Human cell lines mixture (BJ, GM12878, H1 and K562), lysed by Triton-X, HuMix; Human cell lines mixture, lysed by Nuclei EZ Prep, HuMix2; Postnatal day 0 mouse cerebral cortex, P0-brain; Adult mouse cerebral cortex, Ad-brain. d, Histogram showing the numbers of UMIs and genes captured by SNARE-seq expression profiles. e, Histogram showing the number of UMIs and genes detected per nucleus with different single-cell/nucleus RNA-seq methods. The UMI count matrices of published reports were downloaded from GEO (snDrop, GSE97942; SPLiT-seq, GSE110823; sci-CAR, GSE117089). Adult human brain cortex, Brain (H); Postnatal day 2 mouse cerebral cortex, Brain (M).
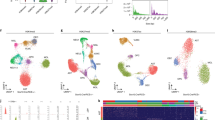
Supplementary Figure 3 SNARE-seq identified cell types within a human cell line mixture (n=1,047).
a, Feature plot showing the marker gene expression of individual cell lines within each cluster. b, Biplot showing the contribution of accessible peak topics (n=11) identified by cisTopic in classifying cell types with chromatin data. c, Dot plot showing the expression of transcription factors (TF) in individual clusters. The size of the dot represents the percentage of nuclei within a cell type expressing the transcription factor and the color indicates the average expression level. d, Motif analysis identified the level of significance (in p-value) of transcription factor binding within differential accessible peak topics (n=404,665 fragments) as mentioned above. One-tailed Fisher’s exact test was used to calculate significance, and Bonferroni correction was made for multiple testing. p-value of marker TF for each cell type is colored in red.
Supplementary Figure 4 Comparison of SNARE-seq dual-omics assay (n=1,043) with single-omic expression (snDrop-seq, n=591) and chromatin (chromatin only, n=494) methods.
a, Clustering of snDrop-seq and SNARE-seq combined expression profiles of human cell line mixture. Cells were labeled by cell type (left) or method (right). b, Clustering of SNARE-seq chromatin profiles (dual or chromatin-only assay) of human cell line mixture. Cells were labeled by cell type (left) or method (right). c, Distribution of transcripts and accessible chromatin peaks detected by SNARE-seq method in individual cell types d, Pearson correlation of gene expression (n=34,828 genes) and chromatin profiles (n=309,891 genomic regions) between dual- and single-omic assays. Aggregated transcript reads and chromatin reads were log10 normalized. e, Distribution of transcripts and chromatin peaks detected by dual- and single-omic assays. The median numbers of transcripts detected by snDrop-seq and SNARE-seq are 1747 and 1159 respectively and the median number of chromatin peaks detected by SNARE-seq single- and dual-omic assay are 2254 and 1960 respectively. In box plots, center lines indicate the median, box limits correspond to the first and third quartiles and whiskers indicate 1.5x interquartile range. f, Species-mixing experiment showing the transcript and chromatin reads detected by SNARE-seq and proportion of human reads in each barcodes.
Supplementary Figure 5 Reproducibility of SNARE-seq (n=5 replicates).
a, Pair-wise correlation of gene-expression profiles between individual replicates of postnatal day 0 sample. Aggregated transcript reads were log10 normalized. b, Pair-wise correlation of chromatin accessibility profiles between individual replicates. Aggregated genome coverage was log10 normalized. c, Proportion of sequencing reads mapped to different genomic features. Top, mapping of reference expression reads, chromatin reads and accessible peaks. Bottom, mapping of SNARE-seq expression reads, chromatin reads and accessible peaks of mouse cerebral cortex data. For this analysis, total expression reads of snDrop-seq and SNARE-seq are 32,059,445 and 8,238,261, respectively. Total chromatin reads and peaks called are 180,548,727 and 140,102, 428,942,515 and 175,298 for snATAC and SNARE-seq, respectively.
Supplementary Figure 6 Robustness of SNARE-seq.
a, Barplot showing the numbers of nuclei recovered for each cell type. UMAP projection of mouse cerebral cortex expression data (n=5,081) as in Fig. 2a showing batch identity (b), and UMI read depth (c). UMAP projection of chromatin accessibility data (n=5,081) as in Fig. 2c showing batch identity (d), and peak read depth (e).
Supplementary Figure 7 Neonatal mouse cerebral cortex SNARE-seq profiles are correlated with published expression and chromatin data.
a, Pearson correlation heatmap of mouse cerebral cortex cell types identified with SNARE-seq expression data (n=4,768) compared with previously identified cell types using SPLiT-seq (n=28,384). b, Intra-assay pair-wise Pearson correlation heatmap of cell types identified with SNARE-seq expression data (n=5,081). c, Intra-assay pair-wise Pearson correlation heatmap of cell types identified with SPLiT-seq expression data (n=28,384). d, Proportion of nuclei in different cell cycle phases showing cell cycle exit of late intermediate progenitor cells. e, Representative Venn diagram showing the number of overlap of common peaks between bulk ATAC-seq (ENCODE) data and SNARE-seq chromatin profiles of mouse brain cortex. f, Aggregated SNARE-seq chromatin profiles (bottom two tracks) agree with bulk ATAC-seq (ENCODE, top two tracks) and are consistent between independent experiments.
Supplementary Figure 8 SNARE-seq expression data identified cell type-specific markers in mouse neonatal cerebral cortex.
a, Dot plot showing the expression of known and novel marker genes (Supplementary Table 1) in each cell type. b, RNA in situ hybridization (ISH) stains (Allen Human Brain) of postnatal (day 4) mouse cerebral cortex showing layer-specific marker gene expression.
Supplementary Figure 9 SNARE-seq expression data identified cell type-specific markers in mouse neonatal cerebral cortex.
a, Dot plot showing the expression of known and novel marker genes (Supplementary Table 1) in each cell type. b, RNA in situ hybridization (ISH) stains (Allen Human Brain) of postnatal (day 4) mouse cerebral cortex showing layer-specific marker gene expression.
Supplementary Figure 10 SNARE-seq links the promoter accessibility with expression level of cell type-specific genes.
a, Top, dot plot showing the differential expression of genes in each cell type. The size of the dot represents the percentage of nuclei within a cell type expressing the gene and the color depth indicates the average expression level. Bottom, dot plot showing the promoter accessibility of markers in each cell type. The size of the dot represents the percentage of nuclei within a cell type that is accessible in promoter regions of corresponding genes and the color depth indicates the average accessibility level. b, Pearson correlation of promoter accessibility and expression levels (n=128 genes) across all cell types for the lineage-specific genes.
Supplementary Figure 11 SNARE-seq chromatin data (n=5,081) from neonatal mouse cerebral cortex uncovers lineage-specific regulatory information.
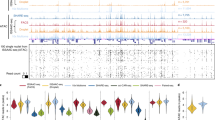
Gene ontology of top biological process was analyzed using a binomial test over 1 kb upstream and 1 kb downstream, up to 500-kb max extension of differential accessible genomic regions in GREAT and transcription factor motifs were discovered using one-tailed Fisher’s exact test and Bonferroni correction in HOMER for each cell type. Interesting examples are shaded in gray.
Supplementary Figure 12 Pseudotime analysis reveals dynamics of gene expression in early neurogenesis.
a, Heatmap of selected genes involved in neurogenesis showing the expression changes along developmental trajectory. b, Promoter accessibility and gene expression dynamics showing mostly similar directional changes along pseudotime. c, Relative expression of gene markers identified by the pseudotime trajectory analysis. The relative expression is calculated by first model read count with the negative binomial and then normalize with cell size factor estimated by estimateSizeFactors function in Monocle. Smoothed pseudotime-dependent gene expression curve as shown in Figure 2g is colored in red. d, Histogram showing the percentage of cells that had accessible promoters calculated by aggregating chromatin signals into bins (bins=30) along pseudotime, and smoothed curve as shown in Figure 2g is colored in orange.
Supplementary Figure 13 Sensitivity of SNARE-seq chromatin data.
a, Total called peaks, recovered promoter regions, median numbers of accessible sites detected per nuclei and total differential accessible sites across all cell types after downsampling raw reads 20-, 15-, 10- and 5-folds. Each dot represents a random downsampling test. Sci-CAR data have a median of ~700 accessible sites/nucleus (indicated by the dash line), which is close to the 15x down-sampled SNARE-seq data. b, Representative UMAP projection of cisTopic clustering result of SNARE-seq chromatin data (n=5,081) at varying down-sampled depths. Cells are labeled with the same color codes for cell types identified by the linked expression data and those discernible clusters are labeled with cluster identities showing on the right.
Supplementary Figure 14 The SNARE-seq profiles of adult mouse cerebral cortex are correlated with published gene expression and chromatin accessibility data.
a, The expression level of marker genes (Supplementary Table 1) for each cell type in adult mouse cerebral cortex. b, Pearson correlation heatmap of mouse cerebral cortex cell types identified with SNARE-seq expression data (n=9,119) compared with previously identified cell types using DroNc-seq (n=4,596). c, Intra-assay pair-wise pearson correlation heatmap of cell types identified with SNARE-seq expression data. d, Intra-assay pair-wise pearson correlation heatmap of cell types identified with DroNc-seq expression data. e, Pair-wise correlation of chromatin accessibility profiles between adult mouse frontal cortex replicates (ENCODE, n=2) and SNARE-seq replicates (n=2). Aggregated genome coverage was log10 normalized. f, In silico downsampling showing the change of genes and UMIs detected by SNARE-seq expression on different sequencing depth.
Supplementary Figure 15 SNARE-seq chromatin data (n=10,309) reveals lineage-specific regulatory information in adult mouse cerebral cortex.
Gene ontology of top biological process (binomial test over 1 kb upstream and 1 kb downstream, up to 500-kb max extension of differential accessible genomic regions in GREAT) and transcription factor motifs analysis (using one-tailed Fisher’s exact test and Bonferroni correction in HOMER) of each cell type in adult mouse cerebral cortex.
Supplementary information
Supplementary Information
Supplementary Figs. 1–15
Supplementary Table 1
Differentially expressed genes between all clusters identified by SNARE-seq expression data.
Supplementary Table 2
Differentially accessible sites between all P0 cerebral cortex clusters identified by SNARE-seq chromatin data.
Supplementary Table 3
Differentially accessible sites between all adult cerebral cortex clusters identified by SNARE-seq chromatin data.
Supplementary Table 4
Primer list.
Rights and permissions
About this article
Cite this article
Chen, S., Lake, B.B. & Zhang, K. High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nat Biotechnol 37, 1452–1457 (2019). https://doi.org/10.1038/s41587-019-0290-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-019-0290-0
This article is cited by
-
InClust+: the deep generative framework with mask modules for multimodal data integration, imputation, and cross-modal generation
BMC Bioinformatics (2024)
-
Decoding leukemia at the single-cell level: clonal architecture, classification, microenvironment, and drug resistance
Experimental Hematology & Oncology (2024)
-
Mosaic integration and knowledge transfer of single-cell multimodal data with MIDAS
Nature Biotechnology (2024)
-
Dictionary learning for integrative, multimodal and scalable single-cell analysis
Nature Biotechnology (2024)
-
Modelling and targeting mechanical forces in organ fibrosis
Nature Reviews Bioengineering (2024)