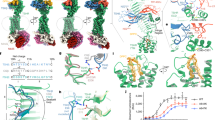
Abstract
Thyroid-stimulating hormone (TSH), through activation of its G-protein-coupled thyrotropin receptor (TSHR), controls the synthesis of thyroid hormone—an essential metabolic hormone1,2,3. Aberrant signalling of TSHR by autoantibodies causes Graves’ disease (hyperthyroidism) and hypothyroidism, both of which affect millions of patients worldwide4. Here we report the active structures of TSHR with TSH and the activating autoantibody M225, both bound to the allosteric agonist ML-1096, as well as an inactivated TSHR structure with the inhibitory antibody K1-707. Both TSH and M22 push the extracellular domain (ECD) of TSHR into an upright active conformation. By contrast, K1-70 blocks TSH binding and cannot push the ECD into the upright conformation. Comparisons of the active and inactivated structures of TSHR with those of the luteinizing hormone/choriogonadotropin receptor (LHCGR) reveal a universal activation mechanism of glycoprotein hormone receptors, in which a conserved ten-residue fragment (P10) from the hinge C-terminal loop mediates ECD interactions with the TSHR transmembrane domain8. One notable feature is that there are more than 15 cholesterols surrounding TSHR, supporting its preferential location in lipid rafts9. These structures also highlight a similar ECD-push mechanism for TSH and autoantibody M22 to activate TSHR, therefore providing the molecular basis for Graves’ disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
10 October 2022
In the version of this article initially published, the Peer review information section did not thank Patrick Scheerer and Gunnar Kleinau for their contributions to the peer review of this article, which is now shown in the HTML and PDF versions of the article.
References
Tuncel, M. Thyroid stimulating hormone receptor. Mol. Imaging Radionucl. Ther. 26, 87–91 (2017).
Allgeier, A. et al. The human thyrotropin receptor activates G-proteins Gs and Gq/11. J. Biol. Chem. 269, 13733–13735 (1994).
Feldt-Rasmussen, U., Effraimidis, G. & Klose, M. The hypothalamus-pituitary-thyroid (HPT)-axis and its role in physiology and pathophysiology of other hypothalamus-pituitary functions. Mol. Cell. Endocrinol. 525, 111173 (2021).
Taylor, P. N. et al. Global epidemiology of hyperthyroidism and hypothyroidism. Nat. Rev. Endocrinol. 14, 301–316 (2018).
Sanders, J. et al. Human monoclonal thyroid stimulating autoantibody. Lancet 362, 126–128 (2003).
Neumann, S. et al. Small-molecule agonists for the thyrotropin receptor stimulate thyroid function in human thyrocytes and mice. Proc. Natl Acad. Sci. USA 106, 12471–12476 (2009).
Evans, M. et al. Monoclonal autoantibodies to the TSH receptor, one with stimulating activity and one with blocking activity, obtained from the same blood sample. Clin. Endocrinol. 73, 404–412 (2010).
Duan, J. et al. Structures of full-length glycoprotein hormone receptor signalling complexes. Nature 598, 688–692 (2021).
Latif, R., Ando, T., Daniel, S. & Davies, T. F. Localization and regulation of thyrotropin receptors within lipid rafts. Endocrinology 144, 4725–4728 (2003).
Fan, Q. R. & Hendrickson, W. A. Structural biology of glycoprotein hormones and their receptors. Endocrine 26, 179–188 (2005).
Jiang, X. et al. Structure of follicle-stimulating hormone in complex with the entire ectodomain of its receptor. Proc. Natl Acad. Sci. USA 109, 12491–12496 (2012).
Kleinau, G. et al. Defining structural and functional dimensions of the extracellular thyrotropin receptor region. J. Biol. Chem. 286, 22622–22631 (2011).
Rapoport, B. & McLachlan, S. M. TSH receptor cleavage into subunits and shedding of the A-subunit; a molecular and clinical perspective. Endocr. Rev. 37, 114–134 (2016).
Couet, J. et al. Cell surface protein disulfide-isomerase is involved in the shedding of human thyrotropin receptor ectodomain. Biochemistry 35, 14800–14805 (1996).
Morshed, S. A. & Davies, T. F. Graves’ disease mechanisms: the role of stimulating, blocking, and cleavage region TSH receptor antibodies. Horm. Metab. Res. 47, 727–734 (2015).
Couet, J. et al. Shedding of human thyrotropin receptor ectodomain. Involvement of a matrix metalloprotease. J. Biol. Chem. 271, 4545–4552 (1996).
Kopp, P. The TSH receptor and its role in thyroid disease. Cell. Mol. Life Sci. 58, 1301–1322 (2001).
Krieger, C. C., Neumann, S. & Gershengorn, M. C. Is there evidence for IGF1R-stimulating Abs in Graves’ orbitopathy pathogenesis? Int. J. Mol. Sci. 21, 6561 (2020).
Dechairo, B. M. et al. Association of the TSHR gene with Graves’ disease: the first disease specific locus. Eur. J. Hum. Genet. 13, 1223–1230 (2005).
Neumann, S. & Gershengorn, M. C. Small molecule TSHR agonists and antagonists. Ann. Endocrinol. 72, 74–76 (2011).
Chazenbalk, G. D. et al. Evidence that the thyrotropin receptor ectodomain contains not one, but two, cleavage sites. Endocrinology 138, 2893–2899 (1997).
Chen, C. R., Salazar, L. M., McLachlan, S. M. & Rapoport, B. Deleting the redundant TSH receptor C-peptide region permits generation of the conformationally intact extracellular domain by insect cells. Endocrinology 156, 2732–2738 (2015).
Ozcabi, B. et al. Testotoxicosis: report of two cases, one with a novel mutation in LHCGR gene. J. Clin. Res. Pediatr. Endocrinol. 7, 242–248 (2015).
Kopp, P. et al. Congenital hyperthyroidism caused by a solitary toxic adenoma harboring a novel somatic mutation (serine281–>isoleucine) in the extracellular domain of the thyrotropin receptor. J. Clin. Invest. 100, 1634–1639 (1997).
Rasmussen, S. G. et al. Crystal structure of the β2 adrenergic receptor–Gs protein complex. Nature 477, 549–555 (2011).
Smits, G. et al. Glycoprotein hormone receptors: determinants in leucine-rich repeats responsible for ligand specificity. EMBO J. 22, 2692–2703 (2003).
Pierce, J. G. & Parsons, T. F. Glycoprotein hormones: structure and function. Annu. Rev. Biochem. 50, 465–495 (1981).
Fox, K. M., Dias, J. A. & Van Roey, P. Three-dimensional structure of human follicle-stimulating hormone. Mol. Endocrinol. 15, 378–389 (2001).
Lapthorn, A. J. et al. Crystal structure of human chorionic gonadotropin. Nature 369, 455–461 (1994).
Grossmann, M., Weintraub, B. D. & Szkudlinski, M. W. Novel insights into the molecular mechanisms of human thyrotropin action: structural, physiological, and therapeutic implications for the glycoprotein hormone family. Endocr. Rev. 18, 476–501 (1997).
Mueller, S., Jaeschke, H., Gunther, R. & Paschke, R. The hinge region: an important receptor component for GPHR function. Trends Endocrinol. Metab. 21, 111–122 (2010).
Bruser, A. et al. The activation mechanism of glycoprotein hormone receptors with implications in the cause and therapy of endocrine diseases. J. Biol. Chem. 291, 508–520 (2016).
van Koppen, C. J. et al. Mechanism of action of a nanomolar potent, allosteric antagonist of the thyroid-stimulating hormone receptor. Br. J. Pharmacol. 165, 2314–2324 (2012).
Sanders, P. et al. Crystal structure of the TSH receptor (TSHR) bound to a blocking-type TSHR autoantibody. J. Mol. Endocrinol. 46, 81–99 (2011).
Vlaeminck-Guillem, V., Ho, S. C., Rodien, P., Vassart, G. & Costagliola, S. Activation of the cAMP pathway by the TSH receptor involves switching of the ectodomain from a tethered inverse agonist to an agonist. Mol. Endocrinol. 16, 736–746 (2002).
Punjani, A. & Fleet, D. J. 3D variability analysis: resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol. 213, 107702 (2021).
Sanders, J. et al. Crystal structure of the TSH receptor in complex with a thyroid-stimulating autoantibody. Thyroid 17, 395–410 (2007).
Neumann, S. et al. An enantiomer of an oral small-molecule TSH receptor agonist exhibits improved pharmacologic properties. Front. Endocrinol. 7, 105 (2016).
Jaeschke, H. et al. An aromatic environment in the vicinity of serine 281 is a structural requirement for thyrotropin receptor function. Endocrinology 147, 1753–1760 (2006).
Kleinau, G. et al. Identification of a novel epitope in the thyroid-stimulating hormone receptor ectodomain acting as intramolecular signaling interface. J. Biol. Chem. 279, 51590–51600 (2004).
Kleinau, G. & Krause, G. Thyrotropin and homologous glycoprotein hormone receptors: structural and functional aspects of extracellular signaling mechanisms. Endocr. Rev. 30, 133–151 (2009).
Schulze, A. et al. The intramolecular agonist is obligate for activation of glycoprotein hormone receptors. FASEB J. 34, 11243–11256 (2020).
Ballesteros, J. A. & Weinstein, H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 25, 366–428 (1995).
Weis, W. I. & Kobilka, B. K. The molecular basis of G protein-coupled receptor activation. Annu. Rev. Biochem. 87, 897–919 (2018).
Katritch, V. et al. Allosteric sodium in class A GPCR signaling. Trends Biochem. Sci. 39, 233–244 (2014).
Kosugi, S., Shenker, A. & Mori, T. Constitutive activation of cyclic AMP but not phosphatidylinositol signaling caused by four mutations in the 6th transmembrane helix of the human thyrotropin receptor. FEBS Lett. 356, 291–294 (1994).
Urizar, E. et al. An activation switch in the rhodopsin family of G protein-coupled receptors: the thyrotropin receptor. J. Biol. Chem. 280, 17135–17141 (2005).
Roberts, S. A., Moon, J. E., Dauber, A. & Smith, J. R. Novel germline mutation (Leu512Met) in the thyrotropin receptor gene (TSHR) leading to sporadic non-autoimmune hyperthyroidism. J. Pediatr. Endocrinol. Metab. 30, 343–347 (2017).
Neumann, S., Krause, G., Chey, S. & Paschke, R. A free carboxylate oxygen in the side chain of position 674 in transmembrane domain 7 is necessary for TSH receptor activation. Mol. Endocrinol. 15, 1294–1305 (2001).
Carpenter, B., Nehme, R., Warne, T., Leslie, A. G. & Tate, C. G. Structure of the adenosine A2A receptor bound to an engineered G protein. Nature 536, 104–107 (2016).
Liang, Y. L. et al. Dominant negative G proteins enhance formation and purification of agonist-GPCR-G protein complexes for structure determination. ACS Pharmacol. Transl. Sci. 1, 12–20 (2018).
Maeda, S., Qu, Q., Robertson, M. J., Skiniotis, G. & Kobilka, B. K. Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes. Science 364, 552–557 (2019).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D 74, 519–530 (2018).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2020).
Lomize, M. A., Pogozheva, I. D., Joo, H., Mosberg, H. I. & Lomize, A. L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 (2012).
Jo, S. et al. CHARMM-GUI 10 years for biomolecular modeling and simulation. J. Comput. Chem. 38, 1114–1124 (2017).
Pascal, B. D. et al. HDX workbench: software for the analysis of H/D exchange MS data. J. Am. Soc. Mass. Spectrom. 23, 1512–1521 (2012).
Acknowledgements
The cryo-EM data were collected at the Cryo-Electron Microscopy Research Center and Advanced Center for Electron Microscopy, Shanghai Institute of Materia Medica (SIMM). We thank the staff at the SIMM Cryo-Electron Microscopy Research Center and Advanced Center for Electron Microscopy for their technical support. This work was partially supported by Ministry of Science and Technology (China) grants (2018YFA0507002 to H.E.X.); Shanghai Municipal Science and Technology Major Project (2019SHZDZX02 to H.E.X.); Shanghai Municipal Science and Technology Major Project (to H.E.X.); CAS Strategic Priority Research Program (XDB37030103 to H.E.X.); the National Natural Science Foundation of China (32130022 to H.E.X., 32171187 to Y. Jiang and 82121005 to H.E.X. and Y. Jiang); CAMS Innovation Fund for Medical Sciences (2021-I2M-1-003 to S.Z.); CAMS Innovation Fund for Medical Sciences (2021-CAMS-JZ004 to S.Z.); Tsinghua University-Peking University Center for Life Sciences (045-61020100121 to S.Z.); National Science & Technology Major Project ‘Key New Drug Creation and Manufacturing Program’ of China (2018ZX09711002 to H.J.); Science and Technology Commission of Shanghai Municipal (20431900100 to H.J.); and the Jack Ma Foundation (2020-CMKYGG-05 to H.J.).
Author information
Authors and Affiliations
Contributions
J.D. designed the expression constructs, purified the TSHR proteins, prepared the final samples for HDX experiments and negative stain, performed cryo-EM grid preparation and data collection, conducted functional studies, and participated in map calculations, figure and manuscript preparation. P.X. performed cryo-EM data calculations, model building and participated in figure preparation. X.L. helped to conceive the project, supplied the TSH hormone and Org274179-0. Q.Y. participated in cryo-EM data calculations. Y. Ji participated in functional studies and protein purification. N.S. performed HDX experiments, supervised by J.Z.; X.H. participated in figure preparation. H.J. and X.C. supervised X.H. in figure preparation. Y. Jiang supervised the studies, and participated in manuscript preparation. S.Z. helped to conceive the project and supervised X.L. and Y. Jin. H.E.X. conceived and supervised the project, analysed the structures and wrote the manuscript with inputs from all of the authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Patrick Scheerer and Gunnar Kleinau and the other anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Cryo-EM images and single-particle reconstruction of the TSH–TSHR–Gs complex.
a, Size-exclusion chromatography elution profile and SDS-PAGE of the TSH–TSHR–Gs complex. Red star indicates the monomer peak of the complex. For gel source data, see Supplementary Fig. 1a. Representative Figures from at least three independent experiments were shown. b, Flowchart of cryo-EM data analysis of the TSH–TSHR–Gs complex. c, d, Cryo-EM micrograph (c), and reference-free 2D class averages (d). 14,965 movies of TSH–TSHR–Gs complex were collected using Titan Krios equipped with a K3 Summit direct electron detector. e, Cryo-EM map of the TSH–TSHR–Gs complex coloured by local resolutions from 2.5 Å (blue) to 5.5 Å (red). f, The density map of TSH–TSHR ECD subcomplex from DeepEMhancer analysis. g, The density map of TSH–TSHR ECD subcomplex from local refinement. h, i, The “Gold-standard” Fourier shell correlation (FSC) curve indicates that the overall resolution of the electron density map of the TSH–TSHR–Gs complex is 2.96 Å, and the local resolution of the electron density map of the TSH–TSHR ECD subcomplex is 2.67 Å.
Extended Data Fig. 2 Cryo-EM image density maps with all transmembrane helices, and H8.
a, TSHR TMD density maps in the TSH–TSHR–Gs complex; b, TSHR TMD density maps in the M22ScFv–TSHR–Gs complex. c, Cholesterol and lipid density maps in the M22ScFv–TSHR–Gs complex.
Extended Data Fig. 3 Structural features of TSH and TSHR in the TSH–TSHR–Gs complex.
a, Cryo-EM density (top panel) and ribbon presentation (bottom panel) of cholesterol molecules around the TSHR TMD in the TSH–TSHR–Gs complex. b, Surface representation of TSHα and TSHβ subunits. The three N-linked glycans are shown in sphere. c, Ribbon presentation of the hinge region in TSHR. TSHR hinge region contains two α-helices (termed as hinge helix 1 and 2), LRR12, a short linker fragment (residues 396-404), and the conserved P10 region (residues 405-414). d, Detail interactions between TSHR and TSHβ subunit. e, Structure comparison of active TSHR and active LHCGR TMD in top view (c) and bottom view (d). f, Structure superposition of TSH–TSHR ECD and K1-70–TSHR ECD. The binding interface of TSH overlaps with K1-70ScFv. g, Concentration-response curves for TSHR cAMP accumulation with K1-70ScFv and 1 nm TSH. Data were shown as mean ± S.E.M. from three independent experiments (n = 3), performed in triplicates. The representative concentration-response curves were shown. h, The positively charged pocket in TSH (left panel) and negatively charged hinge helix 1 surface (right panel), which are highlighted in black circles.
Extended Data Fig. 4 Effects of mutations in the cholesterol-binding motifs on the activation potency of TSH and ML-109 to TSHR.
a, Cholesterol binding sites in the TSHR structure. b, The representative concentration-response curves of TSH- and ML-109-induced WT and mutated TSHR activation. For cAMP analysis, data were shown as mean ± S.E.M. from three independent experiments (n = 3), performed in triplicates. Statistical significance of differences between WT and mutants was determined by two-sided one-way ANOVA with Tukey test. UD, undetectable.
Extended Data Fig. 5 Sequence alignment of glycoprotein hormones and related receptors.
a, Sequences alignment of human TSHR, LHCGR and FSHR in the region of the hormone-binding domain. Residues interact with TSH are labelled in light sea green and blue, while residues that determine TSH–TSHR specificity are labelled in light sea green. b, Sequences alignment of human TSHR, LHCGR and FSHR in the region of the P10 fragments. P10 is shown in orange. c, Sequences alignment of human TSH, CG and FSH β subunit. The major interface of TSHβ interacted with TSHR are highlighted in red, while residues that determine TSH–TSHR specificity are labelled in yellow. d, The α-subunit sequence of glycoprotein hormones. Structure resolved N-linked glycans are highlighted with red stars.
Extended Data Fig. 6 Cryo-EM image and single-particle reconstruction of the K1-70ScFv–TSHR complex.
a, Size-exclusion chromatography elution profile and SDS-PAGE of the K1-70ScFv–TSHR complex. Red star indicates the monomer peak of the two complex. For gel source data, see Supplementary Fig. 1c. Representative Figures from at least three independent experiments were shown. b, Flowchart of cryo-EM data analysis of the K1-70ScFv–TSHR complex. c, d, Cryo-EM micrograph (c), and reference-free 2D class averages (d). 5,938 movies of K1-70ScFv–TSHR complex were collected using Titan Krios equipped with a K3 Summit direct electron detector. e, K1-70ScFv–TSHR complex map and model. f, The “Gold-standard” Fourier shell correlation (FSC) curves indicate that the overall resolution of the K1-70ScFv–TSHR complex is 5.46 Å.
Extended Data Fig. 7 Cryo-EM images and single-particle reconstruction of the M22ScFv–TSHR–Gs complex.
a, Size-exclusion chromatography elution profiles and SDS-PAGEs of the M22ScFv–TSHR–Gs complex. Red star indicates the monomer peak of the complex. For gel source data, see Supplementary Fig. 1b. Representative Figures from at least three independent experiments were shown. b, Flowchart of cryo-EM data analysis of the M22ScFv–TSHR–Gs. c, d, Cryo-EM micrograph (c), and reference-free 2D class averages (d). 6,911 movies of M22ScFv–TSHR–Gs complex were collected using Titan Krios equipped with a K3 Summit direct electron detector. e, Cryo-EM map of the M22ScFv–TSHR–Gs complex coloured by local resolutions from 2.5 Å (blue) to 5.0 Å (red). f, Cryo-EM map of the M22ScFv–TSHR ECD subcomplex from local refinement. g, h, The “Gold-standard” Fourier shell correlation (FSC) curves indicate that the overall resolution of the electron density map of the M22ScFv–TSHR–Gs complex is 2.78 Å, and the local resolution of the electron density map of the M22ScFv–TSHR ECD subcomplex is 2.39 Å.
Extended Data Fig. 8 The binding pocket of ML-109 in TSHR and structure comparison of TSHR and LHCGR.
a, Concentration-response curves for ML-109, TSH, and M22ScFv induced TSHR activation. Data were shown as mean ± S.E.M. from five independent experiments, which performed in triplicates. b, c, Structure comparison of ML-109 binding pocket in the TSH–TSHR–Gs complex with Org43553 binding pocket in the CG–LHCGR–Gs complex. d, Concentration-response curves for ML-109 and CG induced WT and mutated LHCGR activation. Data were shown as mean ± S.E.M. from three independent experiments (n = 3), which performed in triplicates. TM6 means swapping of the extracellular portion of LHCGR TM6 (residues 588-596) with the corresponding TSHR TM6 region (residues 643-651). F515L/A349E/TM6 means mutations of F515L and A349E in addition to the above TM6 mutation. The cAMP data was normalized by CG-induced WT receptor within each individual experiment, with the basal activity for WT as 0, while the fitted Emax of WT as 100. UD, undetectable.
Extended Data Fig. 9 A conserved ECD–TMD configuration for TSHR activation.
a, Structure comparison of TSH–TSHR–Gs complex with M22ScFv–Gs complex. The ECD–TMD interface in two structures are shown. P10 in the TSH–TSHR–Gs complex is shown in blue. b, The two conserved disulfide bonds from the TSHR ECD–TMD interface, which are shown in yellow sticks. c, Detail interactions between ECL1 and I281, Y279 residues. d, The EM density of P10 from the TSH–TSHR–Gs complex. The density map is shown at a level of 0.076. e–g, Detail interactions between P10 and TSHR TMD.
Extended Data Fig. 10 TMD configuration of the TSHR structures.
a, Comparison of HDX-MS analysis on inactivated and active TSHR, differential HDX-MS data consolidated are mapped to active TSHR model according to the differential HDX dynamics key. Regions that show increased HDX activity (more disordered) are coloured yellow/red; regions that show decreased HDX activity (more stable) are coloured green/blue. Comparison of K1-70/Org 274179-TSHR with M22ScFv–TSHR–Gs complex (right panel), comparison of K1-70/Org 274179-TSHR with M22ScFv/ML-109–TSHR–Gs complex (left panel). b–e, Structural comparison between inactivated TSHR and active TSHR. Active TSHR structure from TSH–TSHR–Gs complex is shown in orange, active TSHR structure from M22ScFv–TSHR–Gs complex is shown in green. The inactivated TSHR structure is shown in grey. The two residues M6376.48 and D6336.44 are shown in sticks. f, Structural comparisons between β2AR (PDB: 3SN6) and active TSHR. g, Comparison of inactivated TSHR with ML-109 bound active TSHR. ML-109 clashes with extracellular side of TM6 from the inactivated TSHR structure. h, Comparison the ML-109 binding pocket of TSHR with other orthosteric agonist-binding pockets of class A GPCRs. i, Active TSHR TMD residues locate near the D6336.44 are shown in sticks.
Supplementary information
Supplementary Information
Supplementary Figs. 1–4, Supplementary Tables 1–6 and descriptions for Supplementary Videos 1–4.
Supplementary Video 1
3D viability analysis of TSH–TSHR–Gs complex.
Supplementary Video 2
3D viability analysis of TSH–TSHRECD complex.
Supplementary Video 3
3D viability analysis of M22ScFv–TSHR–Gs complex.
Supplementary Video 4
3D viability analysis of M22ScFv–TSHRECD complex. 3DVA of cryo-EM map reveals trace density of the hinge loop extended from the N terminus of LRR12 and P10 region up to TSH, serving the pull to stabilize the ECD into the upright active position. In contrast, interactions between M22ScFv and the hinge region was not observed, thus suggesting that antibody-mediated TSHR activation is not mediated through a pull mechanism as seen TSHR activated by the endogenous hormone TSH.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Duan, J., Xu, P., Luan, X. et al. Hormone- and antibody-mediated activation of the thyrotropin receptor. Nature 609, 854–859 (2022). https://doi.org/10.1038/s41586-022-05173-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-05173-3
This article is cited by
-
Cryo-electron microscopy for GPCR research and drug discovery in endocrinology and metabolism
Nature Reviews Endocrinology (2024)
-
G protein-coupled receptors (GPCRs): advances in structures, mechanisms, and drug discovery
Signal Transduction and Targeted Therapy (2024)
-
Autoimmune diseases: targets, biology, and drug discovery
Acta Pharmacologica Sinica (2024)
-
Structure, function and drug discovery of GPCR signaling
Molecular Biomedicine (2023)
-
Mechanism of hormone and allosteric agonist mediated activation of follicle stimulating hormone receptor
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.