Abstract
Infections of the central nervous system are among the most serious infections1,2, but the mechanisms by which pathogens access the brain remain poorly understood. The model microorganism Listeria monocytogenes (Lm) is a major foodborne pathogen that causes neurolisteriosis, one of the deadliest infections of the central nervous system3,4. Although immunosuppression is a well-established host risk factor for neurolisteriosis3,5, little is known about the bacterial factors that underlie the neuroinvasion of Lm. Here we develop a clinically relevant experimental model of neurolisteriosis, using hypervirulent neuroinvasive strains6 inoculated in a humanized mouse model of infection7, and we show that the bacterial surface protein InlB protects infected monocytes from Fas-mediated cell death by CD8+ T cells in a manner that depends on c-Met, PI3 kinase and FLIP. This blockade of specific anti-Lm cellular immune killing lengthens the lifespan of infected monocytes, and thereby favours the transfer of Lm from infected monocytes to the brain. The intracellular niche that is created by InlB-mediated cell-autonomous immune resistance also promotes Lm faecal shedding, which accounts for the selection of InlB as a core virulence gene of Lm. We have uncovered a specific mechanism by which a bacterial pathogen confers an increased lifespan to the cells it infects by rendering them resistant to cell-mediated immunity. This promotes the persistence of Lm within the host, its dissemination to the central nervous system and its transmission.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The datasets generated and/or analysed during this study are available as Source Data. Source data are provided with this paper.
References
Schuchat, A. et al. Bacterial meningitis in the United States in 1995. N. Engl. J. Med. 337, 970–976 (1997).
van de Beek, D. et al. Clinical features and prognostic factors in adults with bacterial meningitis. N. Engl. J. Med. 351, 1849–1859 (2004).
Charlier, C. et al. Clinical features and prognostic factors of listeriosis: the MONALISA national prospective cohort study. Lancet Infect. Dis. 17, 510–519 (2017).
Mailles, A. & Stahl, J. Infectious encephalitis in France in 2007: a national prospective study. Clin. Infect. Dis. 49, 1838–1847 (2009).
Skogberg, K. et al. Clinical presentation and outcome of listeriosis in patients with and without immunosuppressive therapy. Clin. Infect. Dis. 14, 815–821 (1992).
Maury, M. M. et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 48, 308–313 (2016).
Disson, O. et al. Conjugated action of two species-specific invasion proteins for fetoplacental listeriosis. Nature 455, 1114–1118 (2008).
Drevets, D. A., Jelinek, T. A. & Freitag, N. E. Listeria monocytogenes-infected phagocytes can initiate central nervous system infection in mice. Infect. Immun. 69, 1344–1350 (2001).
Join-Lambert, O. F. et al. Listeria monocytogenes-infected bone marrow myeloid cells promote bacterial invasion of the central nervous system. Cell. Microbiol. 7, 167–180 (2005).
Cantinelli, T. et al. “Epidemic clones” of Listeria monocytogenes are widespread and ancient clonal groups. J. Clin. Microbiol. 51, 3770–3779 (2013).
Bécavin, C. et al. Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD-e highlights genomic variations underlying differences in pathogenicity. mBio 5, e00969-14 (2014).
Pizarro-Cerdá, J., Lecuit, M. & Cossart, P. in Molecular Cellular Microbiology Vol. 31 (eds Sansonetti, P. & Zychlinsky, A.) 161–177 (Academic Press, 2002).
Boring, L. et al. Impaired monocyte migration and reduced type 1 (Th1) cytokine responses in C-C chemokine receptor 2 knockout mice. J. Clin. Invest. 100, 2552–2561 (1997).
Kocks, C. et al. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell 68, 521–531 (1992).
Tilney, L. G. & Portnoy, D. A. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J. Cell Biol. 109, 1597–1608 (1989).
Lecuit, M. et al. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292, 1722–1725 (2001).
Moura, A. et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2, 16185 (2016).
Gaillard, J. L., Jaubert, F. & Berche, P. The inlAB locus mediates the entry of Listeria monocytogenes into hepatocytes in vivo. J. Exp. Med. 183, 359–369 (1996).
Braun, L. et al. InlB: an invasion protein of Listeria monocytogenes with a novel type of surface association. Mol. Microbiol. 25, 285–294 (1997).
Dramsi, S. et al. Entry of Listeria monocytogenes into hepatocytes requires expression of InIB, a surface protein of the internalin multigene family. Mol. Microbiol. 16, 251–261 (1995).
Lane, F. C. & Unanue, E. R. Requirement of thymus (T) lymphocytes for resistance to listeriosis. J. Exp. Med. 135, 1104–1112 (1972).
Schlüter, D. et al. Systemic immunization induces protective CD4+ and CD8+ T cell‐mediated immune responses in murine Listeria monocytogenes meningoencephalitis. Eur. J. Immunol. 25, 2384–2391 (1995).
Khanna, K. M., McNamara, J. T. & Lefrançois, L. In situ imaging of the endogenous CD8 T cell response to infection. Science 318, 116–120 (2007).
Doherty, P. Cell-mediated cytotoxicity. Cell 75, 607–612 (1993).
Muzio, M. et al. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell 85, 817–827 (1996).
Shen, Y., Naujokas, M., Park, M. & Ireton, K. InIB-dependent internalization of Listeria is mediated by the Met receptor tyrosine kinase. Cell 103, 501–510 (2000).
Bierne, H. et al. A role for cofilin and LIM kinase in Listeria-induced phagocytosis. J. Cell Biol. 155, 101 (2001).
Weidner, K. M. et al. Interaction between Gab1 and the c-Met receptor tyrosine kinase is responsible for epithelial morphogenesis. Nature 384, 173–176 (1996).
Bowers, D. C. et al. Scatter factor/hepatocyte growth factor protects against cytotoxic death in human glioblastoma via phosphatidylinositol 3-kinase- and AKT-dependent pathways. Cancer Res. 60, 4277–4283 (2000).
Xiao, G.-H. et al. Anti-apoptotic signaling by hepatocyte growth factor/Met via the phosphatidylinositol 3-kinase/Akt and mitogen-activated protein kinase pathways. Proc. Natl Acad. Sci. USA 98, 247–252 (2001).
Tsai, Y. H. L., Orsi, R. H., Nightingale, K. K. & Wiedmann, M. Listeria monocytogenes internalins are highly diverse and evolved by recombination and positive selection. Infect. Genet. Evol. 6, 378–389 (2006).
Louie, A., Zhang, T., Becattini, S., Waldor, M. K. & Portnoy, D. A. A multiorgan trafficking circuit provides purifying selection of Listeria monocytogenes virulence genes. mBio 10, e02948-19 (2019).
Coureuil, M., Lécuyer, H., Bourdoulous, S. & Nassif, X. A journey into the brain: insight into how bacterial pathogens cross blood–brain barriers. Nat. Rev. Microbiol. 15, 149–159 (2017).
Devraj, G. et al. HIF-1α is involved in blood–brain barrier dysfunction and paracellular migration of bacteria in pneumococcal meningitis. Acta Neuropathol. 140, 183–208 (2020).
Liu, Y., Li, J. Y., Chen, S. T., Huang, H. R. & Cai, H. The rLrp of Mycobacterium tuberculosis inhibits proinflammatory cytokine production and downregulates APC function in mouse macrophages via a TLR2-mediated PI3K/Akt pathway activation-dependent mechanism. Cell. Mol. Immunol. 13, 729–746 (2016).
Quan, J. H. et al. Intracellular networks of the PI3K/AKT and MAPK pathways for regulating Toxoplasma gondii-induced IL-23 and IL-12 production in human THP-1 cells. PLoS One 10, e0141550 (2015).
Klenerman, P. & Hill, A. T cells and viral persistence: lessons from diverse infections. Nat. Immunol. 6, 873–879 (2005).
Protzer, U., Maini, M. K. & Knolle, P. A. Living in the liver: hepatic infections. Nat. Rev. Immunol. 12, 201–213 (2012).
Mackaness, G. B. Cellular resistance to infection. J. Exp. Med. 116, 381–406 (1962).
Pamer, E. G. Immune responses to Listeria monocytogenes. Nat. Rev. Immunol. 4, 812–823 (2004).
Shen, H. et al. Recombinant Listeria monocytogenes as a live vaccine vehicle for the induction of protective anti-viral cell-mediated immunity. Proc. Natl Acad. Sci. USA 92, 3987–3991 (2006).
Spranger, S., Bao, R. & Gajewski, T. F. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature 523, 231–235 (2015).
Tauriello, D. V. F. et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 554, 538–543 (2018).
Ricci, A. et al. Listeria monocytogenes contamination of ready-to-eat foods and the risk for human health in the EU. EFSA J. 16, 5134 (2018).
Maury, M. M. et al. Spontaneous loss of virulence in natural populations of Listeria monocytogenes. Infect. Immun. 85, e00541-17 (2017).
Shinkai, Y. et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 68, 855–867 (1992).
Malissen, M. et al. Altered T cell development in mice with a targeted mutation of the CD3-epsilon gene. EMBO J. 14, 4641–4653 (1995).
Kitamura, D., Roes, J., Kühn, R. & Rajewsky, K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin μ chain gene. Nature 350, 423–426 (1991).
Jung, S. et al. Analysis of fractalkine receptor CX3CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell. Biol. 20, 4106–4114 (2000).
Hameyer, D. et al. Toxicity of ligand-dependent Cre recombinases and generation of a conditional Cre deleter mouse allowing mosaic recombination in peripheral tissues. Physiol. Genomics 31, 32–41 (2007).
Buch, T. et al. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat. Methods 2, 419–426 (2005).
Matsuzawa, A. et al. A new allele of the lpr locus, lprcg, that complements the gld gene in induction of lymphadenopathy in the mouse. J. Exp. Med. 171, 519–531 (1990).
Kägi, D. et al. Cytotoxicity mediated by T cells and natural killer cells is greatly impaired in perforin-deficient mice. Nature 369, 31–37 (1994).
Huang, Q. Q. et al. FLIP: a novel regulator of macrophage differentiation and granulocyte homeostasis. Blood 116, 4968–4977 (2010).
Huh, C.-G. et al. Hepatocyte growth factor/c-met signaling pathway is required for efficient liver regeneration and repair. Proc. Natl Acad. Sci. USA 101, 4477–4482 (2004).
Canli, Ö. et al. Myeloid cell-derived reactive oxygen species induce epithelial mutagenesis. Cancer Cell 32, 869–883 (2017).
Monk, I. R., Gahan, C. G. M. & Hill, C. Tools for functional postgenomic analysis of listeria monocytogenes. Appl. Environ. Microbiol. 74, 3921–3934 (2008).
Arnaud, M., Chastanet, A. & Débarbouillé, M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, Gram-positive bacteria. Appl. Environ. Microbiol. 70, 6887–6891 (2004).
Argov, T., Rabinovich, L., Sigal, N. & Herskovits, A. A. An effective counterselection system for Listeria monocytogenes and its use to characterize the monocin genomic region of strain 10403S. Appl. Environ. Microbiol. 83, e02927-16 (2017).
Balestrino, D. et al. Single-cell techniques using chromosomally tagged fluorescent bacteria to study Listeria monocytogenes infection processes. Appl. Environ. Microbiol. 76, 3625–3636 (2010).
Quereda, J. J. et al. A dual microscopy-based assay to assess Listeria monocytogenes cellular entry and vacuolar escape. Appl. Environ. Microbiol. 82, 211–217 (2016).
Disson, O. et al. Modeling human listeriosis in natural and genetically engineered animals. Nat. Protoc. 4, 799–810 (2009).
Lu, H. et al. Subcutaneous angiotensin II infusion using osmotic pumps induces aortic aneurysms in mice. J. Vis. Exp. 103, e53191 (2015).
Acknowledgements
We thank P. Bousso and A. Fischer for discussions and the members of the Biology of Infection Unit for their support, in particular L. Travier for technical help on brain microscopy and L. Hafner for contributing to data analysis, and the National Reference Center Listeria. We thank the Cytometry and Biomarkers Unit of Technology and Service (CB UTechS), the Center for Animal Resources and Research (C2RA) and D. Ershov and J.-Y. Tinevez from the Image Analysis Hub at Institut Pasteur. We are grateful to G. de Saint Basile and F. Sepuvelda for the Prf1 KO mice, F. Rieux-Laucat for the Faslpr-cg mice, R. Pope for the Flipflox/flox mice, F. Greten for the LysM-CreERT2 mice, A. Eychene for the Metflox/flox mice and J. Pizarro-Cerda and P. Cossart for the pAD β-lactamase plasmid. Figures 1j, 4l and Extended Data Figs. 5l, 9a were created with BioRender. Work in the M.L. laboratory is funded by the Institut Pasteur, Inserm, ERC, ANR, DIM1HEALTH, Labex IBEID (ANR-10-LABX-62-IBEID) and the Fondation Le Roch-Les Mousquetaires. C.M. and M.K. were recipients of the Roux-Cantarini fellowship of the Institut Pasteur. L.H., C.G. and J.G. were supported by the Université de Paris; Y.-H.T. by the Pasteur–Paris University (PPU) International PhD Program under the European Union’s Horizon 2020 research and innovation program, Marie Sklodowska-Curie grant agreement no 665807; and S.L. by FRM (ECO201906009119) and ‘Ecole Doctorale FIRE-Programme Bettencourt’. M.L. is a member of the Institut Universitaire de France.
Author information
Authors and Affiliations
Contributions
M.L. initiated and coordinated the study. C.M., M.K., S.L. and M.L. conceived and designed the experimental strategy. C.M. and M.K. performed in vivo and cell sorting for ex vivo experiments. C.M. performed flow cytometry experiments. S.L. and Y.-H.T. performed in vivo experiments and imaging of CNS tissue samples. J.G. performed in vivo experiments and imaging of infected intestinal tissue. O.D. and M.K. did ex vivo monocyte imaging. C.M., L.H. and C.G. designed and performed qPCR with reverse transcription experiments. C.M. and L.H. performed cloning and mutagenesis. M.K. performed western blotting experiments. C.M., M.K., S.L., J.G., O.D., Y.-H.T. and M.L. analysed the data. C.M., S.L. and M.L. wrote the manuscript, M.K. and O.D. edited it and all authors agreed on its final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Keith Ireton, Junying Yuan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Infected inflammatory monocytes transfer Lm to the brain.
(a) Bacterial load in the spleen and liver 5 days after oral inoculation with CC4-Lm, in mice treated with gentamicin intraperitoneally every day from day 1 post-inoculation, related to Fig. 1b. (b) Bacterial load in organs after iv inoculation with CC4-Lm, immediately followed by intravenous injection of gentamicin, assessing the bactericidal effect of gentamicin on extracellular circulating Lm. (c) Bacterial load in the blood after oral inoculation with CC1/CC4/CC6-Lm, related to Fig. 1a. (d) Repartition of the 3 main infected cell subsets in the blood and spleen after iv inoculation with CC4-Lm. (e) Number of infected inflammatory monocytes in the spleen and liver of B6-WT mice 3 days after iv inoculation with CC4-Lm, showing that monocytes are infected in the spleen by an order of magnitude more than in the liver. (f) Number of inflammatory monocytes in the blood and spleen of B6-WT or Ccr2−/− mice. (g) Number of bacteria in transferred infected monocytes, collected from spleen of mice 3 days after inoculation with CC4, related to Fig. 1f. (h) Schematic pipeline of the transfer experiment in LysM-CreERT2 × iDTR mice. (i) Bacterial load in the spleen, liver and brain of gentamicin- and diphtheria toxin-treated recipient LysM-CreERT2+/− × Rosa26-iDTR+/− and littermate mice, 4 days after injection of infected monocytes collected from infected donor tamoxifen-treated LysM-CreERT2+/− × Rosa26-iDTR+/− or littermate mice. (j, k) Representative fluorescence microscopy images of brain sections with infected inflammatory monocytes adhering to endothelial cells after iv inoculation with 5×105 CFU CC1-Lm. Adhering infected cells are Ly6C+ (j) and Ly6G− (k). (l) Number of infected monocytes adhering to brain blood vessels of mice inoculated with WT-CC4 or CC4ΔinlB with or without perfusion of vasculature. (m) Representative fluorescence microscopy image of a brain section with infected inflammatory monocytes adhering to endothelial cells after iv inoculation with 5×105 CFU CC1-Lm. Arrow: Lm polymerizing actin in a monocyte; arrowheads: infected monocytes. (n) Representative fluorescence microscopy image of splenocytes 5 days after iv inoculation with 104 CFU CC4-Lm, in which intracellular Lm are found polymerizing actin (top insets) or in LAMP-1 positive vacuole (right insets). (o, p) Proportion of Lm detected polymerizing actin in each infected monocyte (o), or overall fraction of Lm associated with actin or LAMP-1 vacuoles (p) in monocytes from the spleen or from the blood vasculature of mice after inoculation with CC4-Lm. Values in (o) are compared with a t-test and proportions in (p) compared with a χ2 test. Data were obtained from two (a, l) or three (b–h, j–k, m–p) and four (i) independent experiments.
Extended Data Fig. 2 The expression level of InlB is a major determinant of Lm neuroinvasiveness, whereas InlA is not involved.
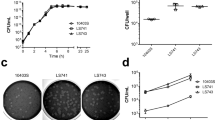
(a) Optical density at 600 nm of indicated bacterial strains measured over time after 1:100 dilution in BHI of an overnight culture. (b, c) Bacterial load after oral inoculation (b) or after iv inoculation (c) with a 1:1 mix of CC4-WT expressing tdTomato or GFP. (d) Bacterial load in brain after iv inoculation with a 1:1 mix of WT and ΔinlA isogenic strains, related to Fig. 2a. (e) Bacterial load in brain after iv inoculation with a 1:1 mix of WT and ΔinlB isogenic strains, related to Fig. 2b. (f, g) Competition indexes (f) and bacterial load (g) in brain after iv inoculation with a 1:1 mix of CC4-WT and either CC4ΔinlA, CC4ΔinlB, CC4ΔinlAB or CC4ΔinlB complemented with inlB (∆inlB + inlB), related to Fig. 2a, b and panels d, e. (h) Bacterial load in brain after oral inoculation with a 1:1 mix of WT strain and ΔinlB isogenic strains, related to Fig. 2c. (i, j) Bacterial load in brain after iv inoculation with 5×103 CFU (i) or oral inoculation with 109 CFU (j) of either CC4-WT or CC4ΔinlB. (k, l) Bacterial load in brain across time after iv inoculation (k) and after oral inoculation (l) with a 1:1 mix of CC4-WT and CC4ΔinlB, related to Fig. 2f, g. (m) Transcription levels of inlA relative to EGDe in mid-log phase in BHI. For CC1/4/6, each dot corresponds to a different clinical isolate and triangles represent the strains used throughout the rest of the study and referred to as CC1, CC4 and CC6, related to panel (o). (n) Transcription levels of inlA relative to EGDe in infected splenocytes 2 days after iv inoculation with 2×105 CFU in mice, related to Fig. 2h. (o) Transcription levels of inlB relative to EGDe in mid-log phase in BHI. Each dot for CC1/4/6 corresponds to a different clinical isolate and triangles point out the strains used throughout the rest of the study and referred to as CC1, CC4 and CC6. (p) Representative Western blot (left) and quantification (right) of InlB expression, normalized to that of EF-Tu, relative to EGDe in mid-log phase in BHI. (q) Bacterial load in brain after inoculation with either WT-EGDe, EGDeΔinlB and WT-CC4. (r) Transcription levels of inlB, relative to EGDe, in infected splenocytes 2 days after iv inoculation with 2×105 CFU of EGDe-WT, CC4-WT and strains complemented with either inlB from EGDe or from CC4. (s) Bacterial load in brain after inoculation with a 1:1 mix of the indicated bacterial strains, related to Fig. 2i. (t) Bacterial load in brain after inoculation with a 1:1 mix of EGDeΔinlB and CC4ΔinlB, related to Fig. 2i. Data were obtained from three independent experiments.
Extended Data Fig. 3 InlB is not involved in Lm invasion of monocytes.
(a, b) Competition indexes (a) and bacterial load (b) in blood after iv inoculation with a 1:1 mix of CC4-WT and CC4ΔinlB. (c) Number of infected monocytes in the spleen after inoculation with CC4-WT or CC4ΔinlB. (d) Bacterial load from 105 sorted monocytes (infected or not) retrieved from mice after inoculation with CC4-WT or CC4ΔinlB. (e) Number of infected monocytes across time in the spleen after inoculation with either CC4-WT or CC4ΔinlB. (f) Number of bacteria per infected monocyte collected from mice after inoculation with either CC4-WT or CC4ΔinlB. (g) Number of infected monocytes in the spleen after inoculation with CC4-WT, CC4ΔinlB or EGDe-WT. (h) Bacterial load in brain of B6-WT or Ccr2−/− mice after inoculation with a 1:1 mix of CC4-WT and CC4ΔinlB, related to Fig. 2l. (i, j) Bacterial load in blood (i) and brain (j) after iv inoculation with 5×105 CFU of either CC4-WT or CC4ΔinlB. (k, l) Competition index (k) and bacterial load (l) in brain 1 day after intracranial inoculation with a 1:1 mix of CC4-WT and CC4ΔinlB. (m-o) Number of infected monocytes (m), percentage of infected monocytes (n) and bacterial load (o) in monocytes 1 h after in vitro infection of primary bone marrow mouse monocytes with WT-Lm or ΔinlB isogenic mutants, at a MOI of 5. Data were obtained from two (k–l), three (a–j) and four (m–o) independent experiments.
Extended Data Fig. 4 Lm neuroinvasion depends on InlB only in the presence of functional CD8+ T cells.
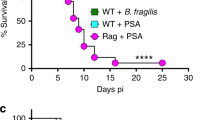
(a) Number of infected monocytes in the blood and spleen of ciclosporin and gentamicin-treated mice after oral inoculation with EGDe. (b, c) Bacterial load in brain (b) and ratio of brain/blood bacterial load (c) in ciclosporin ± gentamicin-treated mice after oral inoculation with EGDe. (d, e) Bacterial load in brain of ciclosporin-treated mice after oral (d) and iv (e) inoculation with a 1:1 mix of EGDe-WT and EGDeΔinlB. (f, g) Bacterial load in brain (f) and in blood (g) of Rag2−/− mice after inoculation with a 1:1 mix of CC4-WT and CC4ΔinlB, related to Fig. 3f. (h) Competition indexes in brain of Rag2−/− mice after inoculation with a 1:1 mix of WT strain and ∆inlB isogenic strains. (i, j) Bacterial load in brain (f) and in blood (g) after inoculation with a 1:1 mix of CC4-WT and CC4∆inlB in control B6 WT mice and in mice lacking functional T (CD3ε−/−), B lymphocytes (muMt−/−) or both (Rag2−/−), related to Fig. 3d. (k) Ratio of brain/blood bacterial loads in B6 WT, Rag2−/−, CD3ε−/− and muMt−/− mice, related to Fig. 3d. (l) Bacterial load in brain of mice after inoculation with a 1:1 mix of CC4-WT and CC4∆inlB after CD8+ T cells depletion, related to Fig. 3e. (m) Representative dot plots (left) and proportion of CD8+ T cells (right) among CD45+ CD3+ cells in the spleen, after CD8+ T cells depletion, related to Fig. 3e. Data were obtained from two (m) or three independent experiments.
Extended Data Fig. 5 InlB does not alter the induction and differentiation of specific anti-Lm CD8+ T cells.
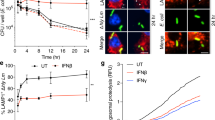
(a, b) Percentage (left) and number (right) of LLO-specific CD8+ T cells in mesenteric lymph nodes (MLN) (a) and spleen (b) of BALB/c mice after iv inoculation with CC4-WT strain or CC4∆inlB. (c, d) Percentage (left) and number (right) of LLO-specific CD8+ T cells in MLN (c) and spleen (d) of iFABP-hEcad mice after oral inoculation with CC4-WT strain or CC4∆inlB. (e–j) Percentage (left) and number (right) of Perforin+ (e), CD69+ (f), Granzyme-B+ (g), CD127+ (h), IFNγ+ (i) and KLRG1+ (j) CD8+ T cells after iv inoculation with CC4-WT or CC4∆inlB. (k) Bacterial load in spleen and liver after oral inoculation with CC4-WT in mice challenged 30 days before with 5x107 CFU of CC4-WT or CC4∆inlB. (l) Schematic pipeline of the cytotoxic lymphocyte (CTL) assay. (m) Level of caspase-3 cleavage of infected spleen monocytes, collected after iv inoculation with CC4-WT or CC4∆inlB, and incubated with CD8+ T cells from similarly infected (WT and ∆inlB) or control (PBS) mice at the indicated effector to target ratio, related to Fig. 3i. Results are normalized to the level of caspase-3 cleavage in absence of CD8+ T cells. (n) Level of caspase-3 cleavage of uninfected spleen monocytes, collected after iv inoculation with CC4-WT or CC4∆inlB, and incubated with CD8+ T cells from similarly infected (WT and ∆inlB) or control (PBS) mice at an effector to target ratio of 5, related to Fig. 3i. Results are normalized to the level of caspase-3 cleavage in absence of CD8+ T cells. Data were obtained from three independent experiments.
Extended Data Fig. 6 Membrane-associated InlB protects infected monocytes from CD8+ T cell-mediated cell death and increases their lifespan, favouring their adhesion to blood brain vessels.
(a) Schematic representation of WT (full length) InlB and its anchored and released variants. (b) Competition indexes in the brain of mice after inoculation with a 1:1 mix of CC4-WT and CC4∆inlB transformed with a plasmid expressing either full-length WT InlB, cell wall-anchored InlB or released InlB. (c) Representative fluorescence microscopy images of centrifugated CC4∆inlB transformed with a plasmid expressing either full length InlB (left panel), anchored InlB (central panel) or released InlB (right panel). Scale bars: 5 μm. (d) Transcription level of inlB in CC4∆inlB transformed with a plasmid expressing InlB variants in mid-log phase in BHI, relative to CC4∆inlB expressing full length InlB. (e) Proportion of infected monocytes containing 1 to 7 bacteria. For monocytes containing more than 1 bacteria, number of GFP- or tdTomato-expressing bacteria in each monocyte is shown. Monocytes were collected 3 days after inoculation with a 1:1 mix of CC4-WT expressing GFP or tdTomato. (f) Level of caspase-3 cleavage of non-infected spleen monocytes, collected from mice infected for 3 days with CC4-WT or CC4∆inlB, incubated ex vivo with FasL, related to Fig. 3m. (g) Percentage of infected spleen monocytes expressing Fas at their surface (left), and the mean fluorescence intensity (MFI) of Fas signal (right), after inoculation with CC4-WT or CC4∆inlB. (h, i) Competition indexes (h) and bacterial load (i) in brain after inoculation with a 1:1 mix of CC4-WT and CC4∆inlB and treatment with caspase-8 inhibitor. (j) Proportion of dye-positive transferred monocytes among infected monocytes in the blood and the spleen after inoculation of B6 WT mice (plain lines) or Rag2−/− (dotted lines) with CC4-WT or CC4∆inlB. Calculated half-lives of infected monocytes are shown in the table. (k) Number of infected monocytes adhering to brain vessels 2 days after inoculation with 5×106 CFU of CC4-WT or CC4∆inlB, expressing tdTomato, of B6 WT mice immunized 2 days before with 103 CFU of CC4-WT expressing GFP. Each dot corresponds to the average number of monocytes counted on two slides (representative median sagittal sections, 40 μm thickness) for one mouse. (l) Number of infected monocytes adhering to brain vessels 2 days after inoculation with 106 CFU of CC4-WT and CC4∆inlB, expressing tdTomato, of Rag2−/− mice immunized 2 days before with 103 CFU of CC4-WT expressing GFP. Each dot corresponds to the average number of monocytes counted on two slides (representative median sagittal sections, 40 μm thickness) for one mouse. Data were obtained from two (k, l) or three independent experiments.
Extended Data Fig. 7 InlB recruits c-Met in LAMP-1+ vacuoles in infected monocytes.
(a) Representative fluorescence microscopy images of spleen monocytes (not permeabilized) collected from tamoxifen-treated LysM-CreERT2 × Metflox/flox (MetLysM∆/∆) and their littermates, showing specific surface expression of c-Met. (b) Percentage of Lm co-localizing with c-Met in vitro in Vero cells 15 min (left), 30 min (middle) and 45 min (right) after infection at MOI 50 with CC4∆inlB expressing either WT InlB, released InlB or cell wall-anchored InlB. (c, d) Representative fluorescence microscopy images of spleen monocytes collected after inoculation with CC4-WT, showing intra-vacuolar Lm surrounded with LAMP-1 (c) and co-localizing with both c-Met and LAMP-1 (d) related to Supplementary Video 4. (e) Representative fluorescence microscopy images of spleen monocytes collected after inoculation with CC4∆inlB, showing no-colocalization with c-Met. (c–e) Maximum intensity projection over a z-stack. (f) Quantification of intracellular Lm co-localizing or not with c-Met and LAMP-1 in infected spleen monocytes collected after inoculation with CC4-WT or CC4∆inlB. Individual cells are plotted in top panel and samples are compared in bottom panel. (g) Representative fluorescence microscopy images of spleen monocytes collected after inoculation with CC4-WT, co-localizing with both c-Met and GAB1 related to Supplementary Video 5. Maximum intensity projection over a z-stack. (h) Bacterial load in brain after inoculation with a 1:1 mix of CC4-WT and CC4∆inlB in mice treated with capmatinib, related to Fig. 4a. (i) Proportion of infected spleen monocytes positive for c-Met signal in flow cytometry after inoculation of MetLysM∆/∆ mice and their littermates with CC4-WT. Data were obtained from three independent experiments (a, h and i) or from three microscopic field of views (f).
Extended Data Fig. 8 InlB-mediated neuroinvasion involves the c-Met–PI3Kα–FLIP pathway in infected monocytes.
(a) Representative fluorescence microscopy images of spleen monocytes collected after inoculation with CC4-WT or CC4∆inlB, showing cytosolic and nuclear phosphorylation of AKT, related to Supplementary Video 6. Images are maximum intensity projection over a z-stack. (b) Proportion of infected spleen monocytes positive for phospho-AKT signal in flow cytometry after inoculation with CC4-WT or CC4∆inlB. (c, d) Competition indexes (c) and bacterial load (d) in brain after inoculation with a 1:1 mix of CC4-WT and CC4∆inlB in mice treated with wortmannin. (e) Competition indexes in brain after inoculation with a 1:1 mix of CC4-WT and CC4∆inlB in mice treated with PI3Kδ inhibitor (IC87114). (f) Bacterial load in the brain after inoculation with a 1:1 mix of CC4-WT and CC4∆inlB in mice treated with BYL-719 or IC87114, related to Fig. 4d and to panel e. (g) Proportion of infected spleen monocytes positive for FLIP signal in flow cytometry after inoculation with CC4-WT, CC4∆inlB or EGDe-WT. (h) Representative dot plot of FLIP expression in infected inflammatory spleen monocytes, after inoculation with CC4-WT or CC4∆inlB, related to Fig. 4e. (i) Proportion of infected spleen monocytes positive for FLIP signal in flow cytometry after inoculation with CC4-WT of MetLysM∆/∆ mice and their littermates after tamoxifen treatment. (j) Level of caspase-3 cleavage of infected spleen monocytes, collected 3 days after inoculation with CC4-WT or CC4∆inlB of tamoxifen-treated Rosa26-CreERT2 × Cflar+/+ (FlipRosa+/+) littermate mice and incubated with CD8+ T cells from similarly infected mice at an effector to target ratio of 5, related to Fig. 4g. (k) Proportion of infected spleen monocytes positive for FLIP signal in flow cytometry, after inoculation with CC4-WT of tamoxifen-treated LysM-CreERT2 × Cflarflox/flox (FlipLysM∆/∆) mice and their littermates (FlipLysM+/+), and after tamoxifen treatment. (l) Bacterial load in tamoxifen-treated (FlipLysM∆/∆), LysM-CreERT2 × Cflarflox/+ (FlipLysM∆/+) and their littermates, after inoculation with a 1:1 mix of CC4-WT and CC4∆inlB and tamoxifen treatment, related to Fig. 4h. Of note, only female mice were used for FlipLysM∆/+, whereas both male and female mice were included for FlipLysM∆/∆and their littermates. (m–o) Proportion of infected spleen monocytes positive for phospho-AKT signal (m), FLIP signal (n) and Zombie signal (o) in flow cytometry, after inoculation with CC4-WT or CC4∆inlB and treatment with HGF. Data were obtained from two (m–o) or three independent experiments.
Extended Data Fig. 9 In the intestinal tract, Lm infects CX3CR1+ macrophages of the lamina propria, in an InlB-dependent manner.
(a) Representation of InlB-activated pathway of infected monocytes survival to Fas-mediated cell death. (b–d) Bacterial load in luminal content of KIE16P (b) and B6 WT (d) or intestinal tissue of B6 WT (c) mice after oral (b) and iv (c, d) inoculation with a 1:1 mix of CC4-WT and CC4∆inlB, related to Fig. 4j, k, Extended Data Fig. 10a, c, e, g. (e) Number of infected CD11b+ CX3CR1+ macrophages of the lamina propria of the small intestine of B6 WT mice in flow cytometry after inoculation with CC4-WT or CC4∆inlB. (f, g) Representative fluorescence microscopy images of infected CX3CR1+ infected macrophages in the colon lamina propria of mice after inoculation with CC4-WT, related to Supplementary Videos 7, 8. Data were obtained from three independent experiments. SI = small intestine.
Extended Data Fig. 10 InlB-mediated Lm gut persistence involves resistance to CD8+ T cells and FLIP expression in myeloid cells.
(a) Competition indexes in intestinal tissue of B6 WT and Rag2−/− mice after inoculation with a 1:1 mix of CC4-WT and CC4∆inlB, related to Extended Data Figs. 9c, d and to panels b–d. (b) Bacterial load in intestinal tissue of Rag2−/− mice after inoculation with a 1:1 mix of CC4-WT and CC4∆inlB, related to a, c, d. (c) Competition indexes in luminal content of B6 WT and Rag2−/− mice after inoculation with a 1:1 mix of CC4-WT and CC4∆inlB, related to Extended Data Fig. 9c, d and to panels a, b, d. (d) Bacterial load in luminal content of Rag2−/− mice after inoculation with a 1:1 mix of CC4-WT and CC4∆inlB, related to panels a–c. (e) Competition indexes in intestinal tissue of mice after inoculation with a 1:1 mix of CC4-WT and CC4∆inlB and treated with an anti-CD8+ T cells antibody, related to Extended Data Fig. 9d and to panels f–h. (f) Bacterial load in intestinal tissue of mice after inoculation with a 1:1 mix of CC4-WT and CC4∆inlB and treated with an anti-CD8+ T cells antibody, related to e–h. (g) Competition indexes in luminal content of mice after inoculation with a 1:1 mix of CC4-WT and CC4∆inlB and treated with an anti-CD8+ T cells antibody, related to Fig. 4j, Extended Data Fig. 9b and to panels e–h. (h) Bacterial load in luminal content of mice after inoculation with a 1:1 mix of CC4-WT and CC4∆inlB and treated with an anti-CD8+ T cells antibody, related to e–g. (i) Competition indexes in intestinal tissue of FlipLysM∆/∆ mice and their littermates after inoculation with a 1:1 mix of CC4-WT and CC4∆inlB, related to Fig. 4k and to panels j–m. (j) Bacterial load in intestinal tissue of littermates of FlipLysM∆/∆ mice after inoculation with a 1:1 mix of CC4-WT and CC4∆inlB, related to Fig. 4k and to panels i, k–m. (k) Competition indexes in luminal content of FlipLysM∆/∆ mice and their littermates after inoculation with a 1:1 mix of CC4-WT and CC4∆inlB, related to Fig. 4k and to panels i, j, l, m. (l) Bacterial load in luminal content of littermates of FlipLysM∆/∆ mice after inoculation with a 1:1 mix of CC4-WT and CC4∆inlB, related to Fig. 4k and to panels i–k, m. (m) Bacterial load in luminal content of FlipLysM∆/∆ mice after inoculation with a 1:1 mix of CC4-WT and CC4∆inlB, related to Fig. 4k and to panels i–l. (n, o) Bacterial load in intestinal tissue (n) and luminal content (o) of mice after inoculation with either CC4-WT or CC4∆inlB and treated with caspase-8 inhibitor, related to Fig. 3k. Data were obtained from three independent experiments. SI = small intestine.
Supplementary information
Supplementary Information
This file contains Supplementary Figs 1–2 and Supplementary Tables 1-7.
41586_2022_4505_MOESM4_ESM.mp4
Supplementary Video 1 Polymerization of actin comet tails by Lm within a monocyte adhering to the blood–brain barrier. CX3CR1GFP/+ KIE16P mice were infected iv with 5x105 CFUs of CC6 for 48 hours. Labels: DNA in blue, CX3CR1 in green, Lm in red, actin in white. 46 optical sections of a 20 µm thick brain sample were imaged with a Zeiss LSM700 confocal microscope. 3D reconstruction was performed using the Arivis Vison 4D software.
41586_2022_4505_MOESM5_ESM.mp4
Supplementary Video 2 Polymerization of actin comet tails by Lm within a monocyte adhering to the blood–brain barrier. KIE16P mice were infected iv 5x105 CFUs of CC4 for 48 hours. Labels: DNA in blue, Ly6C in green, Lm in red, actin in white. Ly6C stains both endothelial cells and monocytes. 100 optical sections of a 20 µm thick brain sample were imaged with a Zeiss LSM900 confocal microscope. 3D reconstruction was performed using the Arivis Vison 4D software.
41586_2022_4505_MOESM6_ESM.mp4
Supplementary Video 3 Lm associated with LAMP-1 and Lm actin comet tails in infected spleen. KIE16P mice were infected iv with 5x103 CFUs of CC4 for 5 days. Labels: DNA in blue, LAMP-1 in red, Lm in green, actin in white. 126 optical sections of a 22.5 µm thick spleen sample were imaged with a Zeiss LSM900 confocal microscope. 3D reconstruction was performed using the Arivis Vison 4D software.
41586_2022_4505_MOESM7_ESM.mp4
Supplementary Video 4 Lm co-localization with c-Met and LAMP-1 in sorted spleen monocytes. KIE16P mice were infected iv with 104 CFUs of CC4 for 4 days. CD45+ CD3- CD19- Ly6G- CD11C- Ly6Chi spleen monocytes were sorted. Labels: Lm in blue, LAMP-1 in green, c-Met in red and DNA in white. 25 optical sections of a 3.8 µm thick spleen sample were imaged with a Zeiss LSM900 confocal microscope. 3D reconstruction was performed using the Arivis Vison 4D software.
41586_2022_4505_MOESM8_ESM.mp4
Supplementary Video 5 Lm co-localization with c-Met and GAB1 in sorted spleen monocytes. KIE16P mice were infected iv with 104 CFUs of CC4 for 4 days. CD45+ CD3- CD19- Ly6G- CD11C- Ly6Chi spleen monocytes were sorted. Labels: Lm in blue, GAB1 in green, c-Met in red and DNA in white. 20 optical sections of a 3.6 µm thick spleen sample were imaged with a Zeiss LSM900 confocal microscope. 3D reconstruction was performed using the Arivis Vison 4D software.
41586_2022_4505_MOESM9_ESM.mp4
Supplementary Video 6 Lm co-localization with c-Met and p-AKT in sorted spleen monocytes. KIE16P mice were infected iv with 104 CFUs of CC4 for 4 days. CD45+ CD3- CD19- Ly6G- CD11C- Ly6Chi spleen monocytes were sorted. Labels: Lm in blue, p-AKT in green, c-Met in red and DNA in white. 37 optical sections of a 6.5 µm thick spleen sample were imaged with a Zeiss LSM900 confocal microscope. 3D reconstruction was performed using the Arivis Vison 4D software.
41586_2022_4505_MOESM10_ESM.mp4
Supplementary Video 7 Infection focus of Lm in the colon. CX3CR1GFP/+ KIE16P mice were infected iv with 5x103 CFUs of CC4 for 5 days. Labels : DNA in blue, CX3CR1 in green, Lm in red, CD11b in white. 16 optical sections of a 5,43 µm thick colon sample were imaged with a Zeiss LSM710 confocal microscope. 3D reconstruction was performed using the Arivis Vison 4D software.
41586_2022_4505_MOESM11_ESM.mp4
Supplementary Video 8 Lm- infected CX3CR1+ cells in the colon. CX3CR1GFP/+ KIE16P mice were infected iv with 5x103 CFUs of CC4 for 5 days. Labels : DNA in blue, CX3CR1 in green, Lm in red, CD8 in white. 33 optical sections of a 11,10 µm thick colon sample were imaged with a Zeiss LSM900 confocal microscope. 3D reconstruction was performed using the Arivis Vison 4D software.
Source data
Rights and permissions
About this article
Cite this article
Maudet, C., Kheloufi, M., Levallois, S. et al. Bacterial inhibition of Fas-mediated killing promotes neuroinvasion and persistence. Nature 603, 900–906 (2022). https://doi.org/10.1038/s41586-022-04505-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-04505-7
This article is cited by
-
Apoptotic cell death in disease—Current understanding of the NCCD 2023
Cell Death & Differentiation (2023)
-
Immunopeptidomics-based design of mRNA vaccine formulations against Listeria monocytogenes
Nature Communications (2022)
-
Listeria hitches a ride
Nature Reviews Microbiology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.