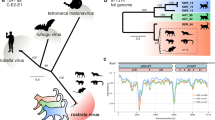
Abstract
Since 1814, when rubella was first described, the origins of the disease and its causative agent, rubella virus (Matonaviridae: Rubivirus), have remained unclear1. Here we describe ruhugu virus and rustrela virus in Africa and Europe, respectively, which are, to our knowledge, the first known relatives of rubella virus. Ruhugu virus, which is the closest relative of rubella virus, was found in apparently healthy cyclops leaf-nosed bats (Hipposideros cyclops) in Uganda. Rustrela virus, which is an outgroup to the clade that comprises rubella and ruhugu viruses, was found in acutely encephalitic placental and marsupial animals at a zoo in Germany and in wild yellow-necked field mice (Apodemus flavicollis) at and near the zoo. Ruhugu and rustrela viruses share an identical genomic architecture with rubella virus2,3. The amino acid sequences of four putative B cell epitopes in the fusion (E1) protein of the rubella, ruhugu and rustrela viruses and two putative T cell epitopes in the capsid protein of the rubella and ruhugu viruses are moderately to highly conserved4,5,6. Modelling of E1 homotrimers in the post-fusion state predicts that ruhugu and rubella viruses have a similar capacity for fusion with the host-cell membrane5. Together, these findings show that some members of the family Matonaviridae can cross substantial barriers between host species and that rubella virus probably has a zoonotic origin. Our findings raise concerns about future zoonotic transmission of rubella-like viruses, but will facilitate comparative studies and animal models of rubella and congenital rubella syndrome.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
17 November 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41586-020-2897-1
References
Lambert, N., Strebel, P., Orenstein, W., Icenogle, J. & Poland, G. A. Rubella. Lancet 385, 2297–2307 (2015).
Zhou, Y., Ushijima, H. & Frey, T. K. Genomic analysis of diverse rubella virus genotypes. J. Gen. Virol. 88, 932–941 (2007).
Chen, J.-P., Strauss, J. H., Strauss, E. G. & Frey, T. K. Characterization of the rubella virus nonstructural protease domain and its cleavage site. J. Virol. 70, 4707–4713 (1996).
Perelygina, L. et al. Infectious vaccine-derived rubella viruses emerge, persist, and evolve in cutaneous granulomas of children with primary immunodeficiencies. PLoS Pathog. 15, e1008080 (2019).
DuBois, R. M. et al. Functional and evolutionary insight from the crystal structure of rubella virus protein E1. Nature 493, 552–556 (2013).
McCarthy, M., Lovett, A., Kerman, R. H., Overstreet, A. & Wolinsky, J. S. Immunodominant T-cell epitopes of rubella virus structural proteins defined by synthetic peptides. J. Virol. 67, 673–681 (1993).
Maton, W. G. Some account of a rash liable to be mistaken for scarlatina. Med. Trans. R. Coll. Physicians 5, 149–165 (1815).
Cooper, L. Z. The history and medical consequences of rubella. Rev. Infect. Dis. 7, S2–S10 (1985).
Gregg, N. M. Congenital cataract following German measles in the mother. Aust. N. Z. J. Ophthalmol. 3, 35–46 (1941).
Parkman, P. D., Buescher, E. L. & Artenstein, M. S. Recovery of rubella virus from army recruits. Proc. Soc. Exp. Biol. Med. 111, 225–230 (1962).
Weller, T. H. & Neva, F. A. Propagation in tissue culture of cytopathic agents from patients with rubella-like illness. Proc. Soc. Exp. Biol. Med. 111, 215–225 (1962).
Swan, C., Tostevin, A. L. & Black, G. H. Final observations on congenital defects in infants following infectious diseases during pregnancy, with special reference to rubella. Med. J. Aust. 2, 889–908 (1946).
Edmunds, W. J., Gay, N. J., Kretzschmar, M., Pebody, R. G. & Wachmann, H. The pre-vaccination epidemiology of measles, mumps and rubella in Europe: implications for modelling studies. Epidemiol. Infect. 125, 635–650 (2000).
Gonzales, J. A. et al. Association of ocular inflammation and rubella virus persistence. JAMA Ophthalmol. 137, 435–438 (2019).
Grant, G. B., Reef, S. E., Patel, M., Knapp, J. K. & Dabbagh, A. Progress in rubella and congenital rubella syndrome control and elimination — worldwide, 2000–2016. MMWR Morb. Mortal. Wkly. Rep. 66, 1256–1260 (2017).
Namuwulya, P. et al. Phylogenetic analysis of rubella viruses identified in Uganda, 2003–2012. J. Med. Virol. 86, 2107–2113 (2014).
Kretsinger, K., Strebel, P., Kezaala, R. & Goodson, J. L. Transitioning lessons learned and assets of the global polio eradication initiative to global and regional measles and rubella elimination. J. Infect. Dis. 216, S308–S315 (2017).
Wolfe, N. D., Dunavan, C. P. & Diamond, J. Origins of major human infectious diseases. Nature 447, 279–283 (2007).
Fahr, J. in Mammals of Africa. Vol. IV: Hedgehogs, Shrews and Bats (eds Happold, M. & Happold, D. C. D.) 380–383 (Bloomsbury, 2013).
Jetz, W., McPherson, J. M. & Guralnick, R. P. Integrating biodiversity distribution knowledge: toward a global map of life. Trends Ecol. Evol. 27, 151–159 (2012).
O’Shea, T. J., Bogan, M. A. & Ellison, L. E. Monitoring Trends in Bat Populations of the United States and Territories: Status of the Science and Recommendations for the Future. Information and Technology Report USGS/BRD/ITR–2003–0003 (US Department of the Interior, US Geological Survey Washington, 2003).
Landau, I. & Chabaud, A.-G. Description de Plasmodium cyclopsi n. sp. parasite du Microchirotère Hipposideros cyclops à Makokou (Gabon). Ann. Parasitol. Hum. Comp. 53, 247–253 (1978).
Schaer, J. et al. High diversity of West African bat malaria parasites and a tight link with rodent Plasmodium taxa. Proc. Natl Acad. Sci. USA 110, 17415–17419 (2013).
Michaux, J. R., Libois, R. & Filippucci, M.-G. So close and so different: comparative phylogeography of two small mammal species, the yellow-necked fieldmouse (Apodemus flavicollis) and the woodmouse (Apodemus sylvaticus) in the Western Palearctic region. Heredity 94, 52–63 (2005).
Labuda, M. et al. Tick-borne encephalitis virus transmission between ticks cofeeding on specific immune natural rodent hosts. Virology 235, 138–143 (1997).
Klempa, B. et al. Complex evolution and epidemiology of Dobrava–Belgrade hantavirus: definition of genotypes and their characteristics. Arch. Virol. 158, 521–529 (2013).
Sibold, C. et al. Dobrava hantavirus causes hemorrhagic fever with renal syndrome in central Europe and is carried by two different Apodemus mice species. J. Med. Virol. 63, 158–167 (2001).
Oktem, I. M. et al. Dobrava–Belgrade virus in Apodemus flavicollis and A. uralensis mice, Turkey. Emerg. Infect. Dis. 20, 121–125 (2014).
Doty, J. B. et al. Isolation and characterization of Akhmeta virus from wild-caught rodents (Apodemus spp.) in Georgia. J. Virol. 93, e00966-19 (2019).
Prpić, J. et al. First evidence of hepatitis E virus infection in a small mammal (yellow-necked mouse) from Croatia. PLoS ONE 14, e0225583 (2019).
Hofmann, J., Renz, M., Meyer, S., von Haeseler, A. & Liebert, U. G. Phylogenetic analysis of rubella virus including new genotype I isolates. Virus Res. 96, 123–128 (2003).
Abernathy, E. et al. Analysis of whole genome sequences of 16 strains of rubella virus from the United States, 1961–2009. Virol. J. 10, 32 (2013).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protocols 10, 845–858 (2015).
Wolinsky, J. S. et al. An antibody- and synthetic peptide-defined rubella virus E1 glycoprotein neutralization domain. J. Virol. 67, 961–968 (1993).
Guy, C., Thiagavel, J., Mideo, N. & Ratcliffe, J. M. Phylogeny matters: revisiting ‘a comparison of bats and rodents as reservoirs of zoonotic viruses’. R. Soc. Open Sci. 6, 181182 (2019).
Luis, A. D. et al. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc. R. Soc. Lond. B 280, 20122753 (2013).
Olival, K. J. et al. Host and viral traits predict zoonotic spillover from mammals. Nature 546, 646–650 (2017).
Frey, T. K. Neurological aspects of rubella virus infection. Intervirology 40, 167–175 (1997).
Bharadwaj, S. D. et al. Acute encephalitis with atypical presentation of rubella in family cluster, India. Emerg. Infect. Dis. 24, 1923–1925 (2018).
Grant, G. B. et al. Accelerating measles and rubella elimination through research and innovation — findings from the Measles & Rubella Initiative research prioritization process, 2016. Vaccine 37, 5754–5761 (2019).
Struhsaker, T. T. Ecology of an African Rain Forest: Logging in Kibale and the Conflict between Conservation and Exploitation (Univ. Press Florida, 1997).
Plumptre, A. J. et al. The biodiversity of the Albertine Rift. Biol. Conserv. 134, 178–194 (2007).
Ulrich, R. G. et al. Network “rodent-borne pathogens” in Germany: longitudinal studies on the geographical distribution and prevalence of hantavirus infections. Parasitol. Res. 103, S121–S129 (2008).
Schlegel, M. et al. Molecular identification of small mammal species using novel cytochrome b gene-derived degenerated primers. Biochem. Genet. 50, 440–447 (2012).
Foley, N. M. et al. How and why overcome the impediments to resolution: lessons from rhinolophid and hipposiderid bats. Mol. Biol. Evol. 32, 313–333 (2015).
Zhao, G. et al. VirusSeeker, a computational pipeline for virus discovery and virome composition analysis. Virology 503, 21–30 (2017).
Bushnell, B. BBMap: a fast, accurate, splice-aware aligner. Version 37.78 https://sourceforge.net/projects/bbmap/ (2014).
Andrews, S. FastQC. A quality control tool for high throughput sequence data. Version 0.11.5 https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2010).
Nurk, S., Meleshko, D., Korobeynikov, A. & Pevzner, P. A. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27, 824–834 (2017).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Huson, D. H. et al. MEGAN community edition — interactive exploration and analysis of large-scale microbiome sequencing data. PLOS Comput. Biol. 12, e1004957 (2016).
Wylezich, C., Papa, A., Beer, M. & Höper, D. A versatile sample processing workflow for metagenomic pathogen detection. Sci. Rep. 8, 13108 (2018).
Scheuch, M., Höper, D. & Beer, M. RIEMS: a software pipeline for sensitive and comprehensive taxonomic classification of reads from metagenomics datasets. BMC Bioinformatics 16, 69 (2015).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Hobman, T. C. & Gillam, S. In vitro and in vivo expression of rubella virus glycoprotein E2: the signal peptide is contained in the C-terminal region of capsid protein. Virology 173, 241–250 (1989).
Gasteiger, E. et al. in The Proteomics Protocols Handbook (ed Walker, J. M.) 571–607 (Humana Press, 2005).
Forth, L. F. & Höper, D. Highly efficient library preparation for ion torrent sequencing using Y-adapters. Biotechniques 67, 229–237 (2019).
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q. & Vinh, L. S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2018).
Mitchell, A. L. et al. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47, D351–D360 (2019).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Rose, A. S. & Hildebrand, P. W. NGL Viewer: a web application for molecular visualization. Nucleic Acids Res. 43, W576–W579 (2015).
Benkert, P., Biasini, M. & Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27, 343–350 (2011).
Korber, B. in Computational Analysis of HIV Molecular Sequences Ch. 4 (eds Rodrigo, A. G. & Learn, G. H.) 55–72 (Kluwer Academic Publishers, 2000).
Leskovec, J. SNAP 2.1. http://snap.stanford.edu/snap-2.1/download.html (2013).
Acknowledgements
We thank D. Hyeroba, K. Swaibu and J. Carag for assistance in the field; C. Langner and the zoo in Germany for assistance with sampling and for implementing timely response strategies; L. Bollinger, J. Wada and D. Rubbenstroth for their help improving the manuscript and figures; G. K. Rice for advice and assistance with bioinformatics scripts; P. Zitzow J. Lorke, S. Schuparis and G. Czerwinski for technical assistance; and C. Jelinek, D. Kaufmann, J. Pöhlig and C. Trapp for help with rodent trapping and dissection. This work was supported in part through US National Institute of Allergy and Infectious Diseases (NIAID) Virology Training Grants T32 AI078985 (to University of Wisconsin-Madison) and GEIS P0062_20_NM_06 (to K.A.B.-L.), and by the Federal Ministry of Education and Research within the research consortium ‘ZooBoCo’ (01KI1722A). This work was also partially supported through the prime contract of Laulima Government Solutions with NIAID under contract no. HHSN272201800013C and Battelle Memorial Institute’s former prime contract with NIAID under contract no. HHSN272200700016I. J.H.K. performed this work as a former employee of Battelle Memorial Institute and a current employee of Tunnell Government Services (TGS), a subcontractor of Laulima Government Solutions under contract no. HHSN272201800013C. Additional support was provided through the German Center for Infection Research (DZIF) TTU ‘Emerging Infections’ (to R.G.U.), and by the University of Wisconsin-Madison Global Health Institute, Institute for Regional and International Studies, and John D. MacArthur Professorship Chair (to T.L.G.). The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies or positions, either expressed or implied, of the US Department of Health and Human Services, Department of the Navy, Department of Defense, US Government, or any of the institutions and companies affiliated with the authors. In no event shall any of these entities have any responsibility or liability for any use, misuse, inability to use, or reliance upon the information contained herein. The US departments do not endorse any products or commercial services mentioned in this publication. K.A.B.-L. is an employee of the US Government. This work was prepared as part of her official duties. Title 17 U.S.C. § 105 provides that ‘Copyright protection under this title is not available for any work of the United States Government.’ Title 17 U.S.C. § 101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person’s official duties. The study protocol was reviewed and approved by the University of Wisconsin-Madison Institutional Animal Care and Use Committee in compliance with all applicable federal regulations governing the protection of animals and research.
Author information
Authors and Affiliations
Contributions
A.J.B., A.C.P., A.E., J.H.K., K.A.B.-L., M.B. and T.L.G. contributed to the study conception and design. A.B., A.J.B., A.C.P., A.E., E.H., G.P., K.A.B.-L., M.B., R.G.U. and T.L.G. contributed to sample and data collection. A.B., A.J.B., A.C.P., A.E., F.P., D.H., E.H., J.H.K., K.A.B.-L., M.B., R.G.U. and T.L.G. contributed to data analyses, interpretation and writing. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Peter Daszak, Fabian Leendertz and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 RNA in situ hybridization of RusV.
a–e, Detection of RusV RNA using in the brain tissues of a donkey (a), red-necked wallaby (b), capybara (c) and yellow-necked field mice (d, e). Chromogenic labelling (fast red) with probes against the NSP-coding region of RusV are visible in neuronal cell bodies (arrow) but not in adjacent glial cells (arrowhead). Scale bars, 50 μm. f, Negative control probe against the bacterial gene dapB, which encodes dihydrodipicolinate reductase. Lack of chromogenic labelling (fast red). Scale bar, 100 µm. All sections were counterstained with Mayer’s haematoxylin. RNAscope results were evaluated on at least three slides per animal, yielding comparable results in all cases. In situ hybridization was performed according to the manufacturer’s instructions, including a positive control probe against peptidylprolyl isomerase B (cyclophilin B) and a negative control probe against dihydrodipicolinate reductase (DapB). Evaluation and interpretation were performed by a board-certified pathologist (DiplECVP) with more than 13 years of experience.
Extended Data Fig. 2 Average substitution rates at non-synonymous and synonymous sites, and the ratio of dN/dS for aligned, concatenated amino acid sequences.
a–c, The average substitution rates at non-synonymous (dN; dashed lines) and synonymous (dS; grey lines) sites, and the ratio of dN/dS (solid lines) for aligned, concatenated amino acid sequences were compared for RuV and RuhV (a), RuV and RusV (b), and RuhV and RusV (c) using sliding windows (100-residue window length, 10 residue steps). Protein domains are labelled on the x axes. MT, methyltransferase; Y, Q and X, domains of unknown function; Pro, protease; Hel, helicase; RdRp, RNA-directed RNA polymerase; NT1, neutralizing epitope 1.
Extended Data Fig. 3 Phylogenetic analyses of the coding sequences of envelope glycoprotein E1, and the helicase and RNA-directed RNA polymerase p90.
a, b, Phylogenetic analyses of the coding sequences (CDS) of the envelope glycoprotein E1 (a) and the helicase and RNA-directed RNA polymerase p90 (b) of RuV, RuhV and RusV, including all sequences obtained in this study (GenBank accession numbers are listed in parentheses). Numbers above branches represent bootstrap values; scale bars indicate amino acid substitutions per site.
Supplementary information
Rights and permissions
About this article
Cite this article
Bennett, A.J., Paskey, A.C., Ebinger, A. et al. Relatives of rubella virus in diverse mammals. Nature 586, 424–428 (2020). https://doi.org/10.1038/s41586-020-2812-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2812-9
This article is cited by
-
Rubella virus genotype 2B endemicity and related utility of serum-based molecular characterization in Uganda
BMC Research Notes (2023)
-
Viruses of Atlantic Bonefish (Albula vulpes) in Florida and the Caribbean show geographic patterns consistent with population declines
Environmental Biology of Fishes (2023)
-
Virale Zoonosen in Deutschland aus der One Health-Perspektive
Bundesgesundheitsblatt - Gesundheitsforschung - Gesundheitsschutz (2023)
-
Mystery of fatal ‘staggering disease’ unravelled: novel rustrela virus causes severe meningoencephalomyelitis in domestic cats
Nature Communications (2023)
-
Relatives of rubella virus
Nature Reviews Microbiology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.