Abstract
Despite its size and rigidity, the cell nucleus can be moved or reorganized by cytoskeletal filaments under various conditions (for example, during viral infection)1,2,3,4,5,6,7,8,9,10,11. Moreover, whereas chromatin organizes into non-random domains12, extensive heterogeneity at the single-cell level13 means that precisely how and why nuclei reorganize remains an area of intense investigation. Here we describe convolutional neural network-based automated cell classification and analysis pipelines, which revealed the extent to which human cytomegalovirus generates nuclear polarity through a virus-assembled microtubule-organizing centre. Acetylation of tubulin enables microtubules emanating from this centre to rotate the nucleus by engaging cytoplasmically exposed dynein-binding domains in the outer nuclear membrane protein nesprin-2G, which polarizes the inner nuclear membrane protein SUN1. This in turn creates intranuclear polarity in emerin, and thereby controls nuclear actin filaments that spatially segregate viral DNA from inactive histones and host DNA, maximizing virus replication. Our findings demonstrate the extent to which viruses can control the nucleus from the cytoplasm.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data generated during this study are included in this published article (and its Supplementary Information files and links provided in the Methods section) or are available upon reasonable request from the corresponding author.
Code availability
All code generated during this study are included in this published article (and its Supplementary Information files and the following links). HCMVcc-CNN: https://github.com/djproc/HCMVcc_MaskRCNN; HCMVcc-Mask-RCNN: https://github.com/djproc/HCMVcc_MaskRCNN; pipelines and quantification: https://github.com/djproc/HCMVcc_pipelines_quant; HCMVcc-CNN: https://hub.docker.com/r/djproc/hcmvcc_cnn; HCMVcc-Mask-RCNN: https://hub.docker.com/r/djproc/hcmvcc_maskrcnn; CNN weights: https://northwestern.box.com/s/g52gt0tqlpgv50hhj3x0dg3t4ljd4he8.
References
Gundersen, G. G. & Worman, H. J. Nuclear positioning. Cell 152, 1376–1389 (2013).
Baarlink, C. et al. A transient pool of nuclear F-actin at mitotic exit controls chromatin organization. Nat. Cell Biol. 19, 1389–1399 (2017).
Christophorou, N. et al. Microtubule-driven nuclear rotations promote meiotic chromosome dynamics. Nat. Cell Biol. 17, 1388–1400 (2015).
Levy, J. R. & Holzbaur, E. L. Dynein drives nuclear rotation during forward progression of motile fibroblasts. J. Cell Sci. 121, 3187–3195 (2008).
Luxton, G. W., Gomes, E. R., Folker, E. S., Vintinner, E. & Gundersen, G. G. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science 329, 956–959 (2010).
Caridi, C. P. et al. Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature 559, 54–60 (2018).
Schrank, B. R. et al. Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature 559, 61–66 (2018).
Wilson, M. H. & Holzbaur, E. L. Opposing microtubule motors drive robust nuclear dynamics in developing muscle cells. J. Cell Sci. 125, 4158–4169 (2012).
Neems, D. S., Garza-Gongora, A. G., Smith, E. D. & Kosak, S. T. Topologically associated domains enriched for lineage-specific genes reveal expression-dependent nuclear topologies during myogenesis. Proc. Natl Acad. Sci. USA 113, E1691–E1700 (2016).
Tsopoulidis, N. et al. T cell receptor-triggered nuclear actin network formation drives CD4+ T cell effector functions. Sci. Immunol. 4, eaav1987 (2019).
Procter, D. J. et al. The HCMV assembly compartment is a dynamic Golgi-derived MTOC that controls nuclear rotation and virus spread. Dev. Cell 45, 83–100.e107 (2018).
Shachar, S. & Misteli, T. Causes and consequences of nuclear gene positioning. J. Cell Sci. 130, 1501–1508 (2017).
Finn, E. H. et al. Extensive heterogeneity and intrinsic variation in spatial genome organization. Cell 176, 1502–1515.e1510 (2019).
Xu, Z. et al. Microtubules acquire resistance from mechanical breakage through intralumenal acetylation. Science 356, 328–332 (2017).
Akella, J. S. et al. MEC-17 is an α-tubulin acetyltransferase. Nature 467, 218–222 (2010).
Zhu, R., Antoku, S. & Gundersen, G. G. Centrifugal displacement of nuclei reveals multiple LINC complex mechanisms for homeostatic nuclear positioning. Curr. Biol. 27, 3097–3110.e3095 (2017).
Buchkovich, N. J., Maguire, T. G. & Alwine, J. C. Role of the endoplasmic reticulum chaperone BiP, SUN domain proteins, and dynein in altering nuclear morphology during human cytomegalovirus infection. J. Virol. 84, 7005–7017 (2010).
Swan, A., Nguyen, T. & Suter, B. Drosophila Lissencephaly-1 functions with Bic-D and dynein in oocyte determination and nuclear positioning. Nat. Cell Biol. 1, 444–449 (1999).
Milbradt, J. et al. Proteomic analysis of the multimeric nuclear egress complex of human cytomegalovirus. Mol. Cell. Proteomics 13, 2132–2146 (2014).
Berk, J. M., Tifft, K. E. & Wilson, K. L. The nuclear envelope LEM-domain protein emerin. Nucleus 4, 298–314 (2013).
Holaska, J. M., Kowalski, A. K. & Wilson, K. L. Emerin caps the pointed end of actin filaments: evidence for an actin cortical network at the nuclear inner membrane. PLoS Biol. 2, E231 (2004).
Baarlink, C., Wang, H. & Grosse, R. Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science 340, 864–867 (2013).
Plessner, M., Melak, M., Chinchilla, P., Baarlink, C. & Grosse, R. Nuclear F-actin formation and reorganization upon cell spreading. J. Biol. Chem. 290, 11209–11216 (2015).
Serebryannyy, L. A. et al. Persistent nuclear actin filaments inhibit transcription by RNA polymerase II. J. Cell Sci. 129, 3412–3425 (2016).
Forest, T., Barnard, S. & Baines, J. D. Active intranuclear movement of herpesvirus capsids. Nat. Cell Biol. 7, 429–431 (2005).
Wilkie, A. R., Lawler, J. L. & Coen, D. M. A role for nuclear F-actin induction in human cytomegalovirus nuclear egress. MBio 7, e01254-16 (2016).
Bosse, J. B. & Enquist, L. W. The diffusive way out: herpesviruses remodel the host nucleus, enabling capsids to access the inner nuclear membrane. Nucleus 7, 13–19 (2016).
Feierbach, B., Piccinotti, S., Bisher, M., Denk, W. & Enquist, L. W. Alpha-herpesvirus infection induces the formation of nuclear actin filaments. PLoS Pathog. 2, e85 (2006).
Chang, L. et al. Herpesviral replication compartments move and coalesce at nuclear speckles to enhance export of viral late mRNA. Proc. Natl Acad. Sci. USA 108, E136–E144 (2011).
Luo, M. H., Rosenke, K., Czornak, K. & Fortunato, E. A. Human cytomegalovirus disrupts both ataxia telangiectasia mutated protein (ATM)- and ATM-Rad3-related kinase-mediated DNA damage responses during lytic infection. J. Virol. 81, 1934–1950 (2007).
Crisp, M. et al. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 172, 41–53 (2006).
Plessner, M., Knerr, J. & Grosse, R. Centrosomal actin assembly is required for proper mitotic spindle formation and chromosome congression. iScience 15, 274–281 (2019).
Belin, B. J., Lee, T. & Mullins, R. D. DNA damage induces nuclear actin filament assembly by Formin -2 and Spire-1/2 that promotes efficient DNA repair. eLife 4, e07735 (2015).
Xu, J. et al. Super-resolution imaging of higher-order chromatin structures at different epigenomic states in single mammalian cells. Cell Rep. 24, 873–882 (2018).
Ovesný, M., Křížek, P., Borkovec, J., Svindrych, Z. & Hagen, G. M. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 30, 2389–2390 (2014).
Chaumeil, J., Micsinai, M. & Skok, J. A. Combined immunofluorescence and DNA FISH on 3D-preserved interphase nuclei to study changes in 3D nuclear organization. J. Vis. Exp. 2013, e50087 (2013).
McQuin, C. et al. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 16, e2005970 (2018).
He, K., Gkioxari, G., Dollár, P. & Girshick, R. Mask R-CNN. Preprint at https://arxiv.org/abs/1703.06870 (2018).
Acknowledgements
We thank E. Murphy, C. O’Connor, G. Gundersen, T. Shenk, J. Holaska, R. Grosse and A. Carpenter for reagents and advice. This work was supported by grants from the National Institutes of Health (NIH) (R01AI141470 and P01GM105536 to D.W.).
Author information
Authors and Affiliations
Contributions
D.J.P. designed the CNN analysis pipeline and performed most experiments. D.J.P. was assisted by C.F. in performing infection analyses including western blotting, immunofluorescence and determining viral titres. D.J.P. and A.G.G.-G. performed DNA-immunoFISH. S.T.K. provided support and advice on genome organization and DNA-immunoFISH. D.J.P. and D.W. designed experiments, analysed data and wrote the manuscript. All authors read and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Robert Grosse and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Tubulin acetylation regulates nuclear rotation and SUN1 polarization during HCMV infection.
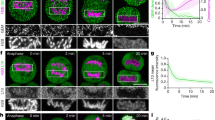
a, b, Representative stills from time lapse imaging and measurements of rotation frequency above or below 180° in uninfected or infected NHDFs expressing GFP-Histone nanobody. Bars represent mean ± SEM, statistics use two-tailed Student’s t-test, n = 281 cells total from 3 independent biological replicates; ****P ≤ 0.0001. Note that nuclear rotation above 180° occurs in approximately 80% of infected cells imaged, while lower levels of rotation occur in the remaining population. Such extensive rotation is extremely rare in uninfected cells. c, d, Expression of a K40R mutant form of tubulin suppresses the formation of acetylated microtubule filaments. Fluorescence intensity of acetylated tubulin is shown in b; All data points are shown within violin plots, statistics use two-tailed Student’s t-test, n = 250 cells total, ****P ≤ 0.0001. Data shown is representative of 3 independent biological replicates. e, f, Expression of a K40R mutant form of tubulin suppresses nuclear rotation. Representative stills from Supplementary Video 2 are shown in e and rotational analyses are shown in f. Rotation frequency above or below 180° is shown in d; bars represent mean ± SEM, statistics use two-tailed Student’s t-test, n = 157 cells total, **P ≤ 0.01 g, Schematic of CNN-based classification and analysis pipeline measuring fluorescence intensities across individual cells in different channels. Output for the assembly compartment (red) next to the nucleus (blue) is illustrated. h, Representative confocal z-section and deconvolved z-section image of SUN1 polarization in HCMV-infected cell. Acetylated microtubules and the assembly compartment (stained with the viral protein gB) are also shown. Data shown is representative of 3 independent biological replicates. i, Spatial distribution and intensity of DNA, gB and SUN2 using CNN. Lines represent mean ± SEM; n ≥ 17,484 cells total from 3 independent biological replicates.
Extended Data Fig. 2 Effects of HCMV infection on SUN1.
a, WB analysis of SUN1 levels over the course of infection with HCMV at MOI 1. Early (IE1/2), intermediate (UL44) and late (pp65, pp28) proteins demonstrate stages of infection at each time point, representative of 3 independent biological replicates. b–d, Neural network-based single cell analysis of SUN1 expression during HCMV infection. b, Illustration of CNN analysis pipeline that classifies cells by the predominant infectious cycle stage identified at each time point. c, Representative examples of uninfected NHDFs or NHDFs at various stages of infection, stained for SUN1, IE1/2 and TGN46. Expression of IE1/2 and gradual remodelling of the Golgi network serve as markers of infection stage. Polarization of SUN1 is seen between 24-72 h.p.i. d, CNN-based classification of cells based on IE1/2 expression levels, filtering out uninfected cells, reveals a gradual expansion of the nucleus and Golgi, characteristic of HCMV infection, occurs concomitantly with a gradual increase in expression and polarization of SUN1 towards the assembly compartment (i-v). Discrete populations of cells are filtered for inclusion in each time point (vii), with cells from other kinetic classes removed from analysis marked in grey (viii-x). Comparing unfiltered (lighter colored violin plots, left segment) versus filtered (dark colored violin plots, right segment) cell populations reveals the power of trained networks to more precisely analyse only infected cells within the population, more clearly revealing the increase in nuclear volume and SUN1 abundance, which peaks at approximately twofold (xi-xiii). Lines represent mean ± SEM; n = 37,800 cells total from 3 independent biological replicates. Violins as in Fig. 4a. e, Mask-RCNN analysis pipeline uses manually annotated masks of the assembly compartment, nucleus and combined (HCMV) to train a Mask-RCNN architecture to classify and segment microscopy images of HCMV infection. Once trained, whole coverslip scanning data sets can be run through the model to perform instantaneous single cell quantification on high-confidence infected cells. This quantification has high spatial awareness and can be used to perform linescans between two specific subcellular compartments (for example, the assembly compartment and nucleus) or to rotate and align nuclei to perform average projections (as in Fig. 1g).
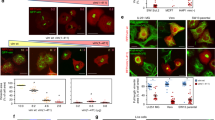
Extended Data Fig. 3 SUN1:Nesprin-2G and the dynein adaptor BICD2 mediate nuclear rotation during HCMV infection.
a, b, Expression of a SUN1 mutant that does not engage Nesprin-2G impairs nuclear rotation in HCMV-infected cells. a, Representative stills from time lapse recordings of NHDFs expressing Tag-GFP2 forms of SUN1 Full Length (FL) or SUN1 lacking the lumenal domain (SUN1ΔLu) that mediates interactions with Nesprin-2G, infected with HCMV-UL99mCherry. Rotation traces from this imaging are shown to the right. Analyses focused on cells expressing intermediate levels of SUN1-GFP constructs as high levels of expression can result in retention of Nesprin-2G in the endoplasmic reticulum (ER). b, Quantification of rotation frequencies above or below 180°; bars represent mean ± SEM, statistics use two-tailed Student’s t-test, n = 138 cells total from 3 independent biological replicates, ***P ≤ 0.001. This data further confirms that interactions with Nesprin-2G are necessary for nuclear rotation to occur. c, Illustration of SUN1:Nesprin-2G interactions with microtubule motors through adaptive domain regions, or SUN2:Nesprin-2G interactions with myosin through calponin homology domains to control nuclear movement. d, Illustration of GFP-Nesprin-2G constructs with calponin homology and/or adaptive domain domains, along with the LEWD > LEAA kinesin-binding mutant. e–h, RNAi-mediated depletion of BICD2 using either of two independent siRNAs suppresses nuclear rotation and SUN1 polarization. e, Western blot analysis of BICD2 expression representative of 3 independent replicates. Arrow points to BICD2, specifically depleted by two independent siRNAs. f, Representative stills and rotational analyses from Supplementary Video 6 showing effects of BICD2 depletion on nuclear rotation. g, Rotation frequency above or below 180° in control or BICD2 depleted cells, bars represent mean ± SEM, statistics use two-tailed Student’s t-test, n = 144 cells total cells from n = 3-4 independent biological replicates, ***P ≤ 0.001. h, Depletion of BICD2 impairs SUN1 polarization. Spatial distribution and intensity of DNA (hoescht), assembly compartment marker (gB), and SUN1 are shown for control and BICD2 depleted cells and are representative of 3 independent biological replicates. For quantification, SUN1 was classed as polarized (green), intermediate (pink) or non-polarized (orange). n = 235 cells total. i–k, Expression of a dominant-negative fragment of BICD2 reduces nuclear rotation and SUN1 polarization. NHDFs expressing TagGFP2 control or TagGFP2-BICD2 N terminus (NT) were infected with HCMV UL99-mCherry. i, Representative still images from time lapse recordings and rotation traces are shown. j, Quantification of rotation frequencies above or below 180°; bars represent mean ± SEM, n = 91 cells total from 2 independent biological replicates. k, Representative images of SUN1 localization in NHDFs expressing TagGFP2 control or TagGFP2-BICD2-NT NHDFs are shown, consistent with 3 independent biological replicates. Quantification of SUN1 polarity categorized as fully polarized, intermediate polarity or not polarized is shown; n = 149 cells total.
Extended Data Fig. 4 Microtubules and SUN1 regulate Emerin polarity and nuclear F-actin.
a, Lamin A/C is downregulated and lacks polarity in HCMV-infected cells. Lines represent mean ± SEM; n = 10,934 cells total from 3 independent biological replicates. b, Depletion of αTAT1, SUN1 or BICD2 inhibits Emerin polarization and causes aberrant F-actin networks. Representative images are shown for each condition, similar to data from 3 independent replicates. c, d, Emerin depletion blocks nuclear F-actin formation. c, WB analysis demonstrating the efficacy of Emerin siRNAs. d, Representative images and quantification of nuclear F-actin (nAC) frequency are shown for each condition, bars represent mean ± SEM, statistics use two-tailed Student’s t-test, n = 401 cells total from 3 independent biological replicates, ***P ≤ 0.001. Fluorescence intensity shows Emerin depletion in cells. e, Emerin depletion does not affect SUN1 polarization. Representative images and quantification of SUN1 polarization is shown for each condition; n = 321 cells. SUN1 was characterized as polarized, intermediate polarity or not polarized. f, Expression of actin-binding mutants of Emerin blocks nuclear F-actin formation but not nuclear rotation. NHDFs expressing nAC-TagGFP2 and mCherry-Emerin wildtype or actin-binding mutants (m151, m175) were infected with HCMV UL99-mCherry. Representative still images and rotation traces from time lapse imaging are shown. Quantification of nuclear rotation frequencies above or below 180° are shown for each condition; the presence of nuclear F-actin was also quantified in the same time lapse images, n = 72 cells total (upper) and n = 79 cells total (lower). Note that in order to image nAC-TagGFP2 cells were infected with HCMV UL99-mCherry. As such, mCherry signal in these images originates from both mCherry-Emerin and the viral UL99-mCherry, showing the cytoplasmic assembly compartment and nuclear rotation in infected cells under all conditions. Data shown is representative of 3 independent replicates.
Extended Data Fig. 5 Nuclear F-actin formation and histone modifications during HCMV infection.
a, γH2AX localizes to nuclear F-actin and polarizes during HCMV infection. Representative images of HCMV-infected cells expressing nAC-TagGFP2 to detect nuclear actin filaments, fixed at the indicated times post-infection and stained for histone γH2AX. Intensity heat maps are shown at the top, illustrating the appearance and gradual polarization of γH2AX foci. In merges, γH2AX foci (purple staining) are observed adjacent to nuclear actin filaments (green) during the rotation phase, and are highly polarized by the time nuclear rotation ceases and actin filaments disassemble. b, Fixed images showing γH2AX and nuclear F-actin induction in nAC-TagGFP2-expressing NHDFs treated with the DNA damage agent, etoposide. c, Still images from Supplementary Video 9 showing the formation of thick nuclear F-actin in response to etoposide. d, Representative examples of the spatial distribution of H3K9me3 foci at early time-points in HCMV infection. Infected cells were identified by staining for IE1/2. Average H3K9me3 fluorescence intensity per cell is shown in violin plots with all data points shown, statistics use two-tailed Student’s t-test n = 8,177 cells total; ****P ≤ 0.0001, ns = not significant. Note that H3K9 trimethylation increases by 24 h.p.i. but is not polarized before the nuclear rotation phase of infection. e, Representative still images from Supplementary Video 10 showing the localization of histones visualized in NHDFs expressing eGFP-Histone nanobody and infected with HCMV UL99-mCherry. Rotation trace is shown to the right. Note that histones are dynamic but are next extensively polarized, in line with fixed images in Fig. 4a. f, Arp2/3 inhibitor, CK-666 blocks nuclear actin filament formation and polarization of histone H3K9me3. Representative images are shown at 48 and 96 h.p.i., the peak and end-point of nuclear rotation and establishment of polarity, respectively. Insets show the localization of H3K9me3 foci near nuclear F-actin at the early stages of rotation when polarity is being established. For all experiments, data shown is representative of 3 independent biological replicates.
Extended Data Fig. 6 Localization of viral genomic DNA and histones in HCMV-infected cells.
a, Representative images of early and mature replication compartments containing viral DNA in HCMV-infected cells, versus uninfected (mock) cells, detected using FISH. DNA-immunoFISH was used to detect the viral immediate early transcription factor, IE1/2 in conjunction with viral DNA. IE1/2 is present in cells containing early, individual replication compartments. As infection progresses, IE1/2 abundance increases and DNA-containing replication compartments amplify and coalesce. b, Representative examples of the relative spatial distribution and fluorescence intensity of histone H3K9me3 foci and viral genomic DNA (gDNA) in HCMV-infected cells at 72 and 96 h.p.i. Note that the bulk of histone H3K9me3 foci are spatially polarized towards the assembly compartment and segregated away from viral gDNA. For all experiments, data shown is representative of 3 independent biological replicates.
Extended Data Fig. 7 d-STORM imaging reveals the segregation of host and viral DNA during HCMV infection.
Differential EdU-labelling strategies enable super-resolution imaging of host and viral chromatin structures and localization. Cartoons to the left illustrate each labelling strategy. Top, Localization of host DNA in uninfected cells was visualized by pulsing with EdU followed by d-STORM imaging. Images representative of 3 independent biological replicates are shown, illustrating how labelled DNA is distributed throughout the nucleus. Middle, Localizaton of host DNA in HCMV-infected cells. To selectively label host DNA during infection, cells were pulsed with EdU which was then removed before infection to prevent incorporation into viral DNA. Representative d-STORM images are shown of labelled host DNA at 72 h.p.i., illustrating its accumulation near the viral assembly compartment. Lower, Localizaton of viral DNA in HCMV-infected cells. To selectively label viral DNA but not host DNA, cells were pulsed with EdU at 72 h.p.i. As infection blocks host DNA synthesis, EdU is only incorporated into viral DNA at this time. Representative d-STORM images are shown of viral DNA at 72 h.p.i., illustrating its accumulation on the opposing side of the nucleus, away from the viral assembly compartment and regions containing host DNA or heterochromatin. This pattern of viral DNA labelling is validated by DNA Immuno-FISH imaging of viral DNA in Extended Data Fig. 6.
Extended Data Fig. 8 BICD2, SUN1 and Emerin are required for the polarization of heterochromatin by HCMV.
a, Depletion of αTAT1, BICD2 or SUN1 reduces H3K9me3 polarization. Representative images are shown. b, Expression of BICD2 dominant-negative (BICD2-NT) or SUN1 lacking its lumenal domain (SUN-ΔLu) that mediates interactions with Nesprin-2G inhibits the polarization of H3K9me3 in HCMV-infected cells. Representative images and quantification of H3K9me3 polarization are shown; n = 171 cells total. c, Depletion of Emerin impairs the polarization of H3K9me3 in HCMV-infected cells. Representative images and quantification of H3K9me3 polarization are shown; n = 490 cells total. For all experiments, data shown is representative of 3 independent biological replicates.
Extended Data Fig. 9 Host factors that control nuclear polarization are required for efficient HCMV replication.
a, b, siRNAs targeting ATAT1, BICD2, SUN1 or Emerin do not affect the accumulation of viral proteins from different kinetic classes, but reduce the production of infectious virus in cells infected at MOI 1. Reductions in viral yields are similar to reductions in DNA fluorescence intensities detected in Fig. 4c. This suggests that nuclear polarization does not regulate viral gene expression, but maximizes viral DNA replication and production of infectious virus. Bars represent mean ± SEM from 3 independent biological replicates, statistics use two-tailed Student’s t-test, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. c, Depletion of ATAT1, BICD2 or SUN1 potently suppresses HCMV spread. Quantification of plaque areas is shown to the right in e; Bars represent mean ± SEM, statistics use two-tailed Student’s t-test, n = 31-81 plaques total from 3 independent biological replicates, *P ≤ 0.05, **P ≤ 0.01. This data suggests that while viral DNA replication and virus yields are reduced by 50% in a single round of infection, this has cumulative effects on the ability of HCMV to spread to other cells. d–f, General inhibition of Arp2/3 activity has broader effects on HCMV infection. d, The Arp2/3 inhibitor CK-666 suppresses viral gDNA polarization and accumulation. Representative images are shown of viral gene expression. gDNA and its localization relative to H3K9me3 foci. e, HCMV spread is suppressed in CK-666-treated, but not inactive control CK-689-treated cells. Plaque areas are shown, bars represent mean ± SEM, statistics use two-tailed Student’s t-test, n = 416 plaques total from 3 independent biological replicates, *P ≤ 0.05, **P ≤ 0.01. f, Arp2/3 inhibition suppresses viral protein accumulation and virus yields in single cycle infections. These data show that inhibition of actin polymerization using Arp2/3 inhibitors not only blocks nuclear polarization but has additional effects on viral gene expression, including modest effects on IE2 expression and more noticeable effects on intermediate and late proteins. This suggests roles for both nuclear and cytoplasmic actin when using broad-spectrum inhibitors over more targeted approaches against nuclear actin alone. Bars represent mean ± SEM from 3 independent biological replicates, statistics use two-tailed Student’s t-test, *P ≤ 0.05, ***P ≤ 0.001. For all experiments, data shown is representative of 3 independent biological replicates.
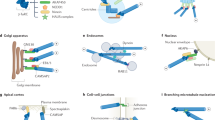
Extended Data Fig. 10 A model for HCMV-induced nuclear polarization.
Top, In uninfected cells chromatin and silenced domains are heterogeneously distributed throughout the nucleus, as discussed in the main text. Middle, Upon HCMV infection, nuclear F-actin is induced and reorganized through the action of acetylated microtubules that exert mechanotransductive pulling forces on Nesprin-2G:SUN1-containing LINC complexes, polarizing them towards the assembly compartment. In doing so, this creates extreme polarity in inner-nuclear Emerin, directing nuclear F-actin organization; enriched red regions represent polarized LINC-Emerin complexes in the nuclear membrane. This extreme polarity draws silenced (H3K9me3) histones and associated host DNA towards this region of the nucleus, through the action of nuclear F-actin networks. As viruses employ a wide range of strategies to prevent chromatinization and silencing of their own DNA, viral gDNA is not drawn to the assembly compartment-proximal sites of H3K9me3 polarization. The polarization of inactive histones and host DNA likely pushes viral DNA to the opposing side of the nucleus, through space-filling. This segregation of viral and host DNA creates an optimal environment for viral DNA replication and production of infectious virus particles. Lower, Polarization of the nucleus fails to occur if key components driving the process are inhibited; if microtubules are not mechanically strengthened through acetylation, if connections between microtubules and nuclear membrane complexes are lost, or if nuclear F-actin is not organized by Emerin. Notably, nuclear F-actin and Emerin do not control nuclear rotation, but cytoplasmic microtubule-derived forces that cause nuclear rotation control Emerin localization, F-actin formation and intranuclear polarity. As such, cytoplasmic forces on the nuclear surface organize nuclear factors to control genetic polarity. Failure to create this polarity results in a suboptimal environment for viral DNA replication.
Supplementary information
Supplementary Figure
Supplementary Figure 1: Raw Blots for Western Blotting images. Cropped regions shown in figures are highlighted.
Video 1
αTAT1 is required for efficient nuclear rotation by HCMV. NHDFs were infected with TB40/E-UL99-eGFP followed by treatment with control or αTAT1 siRNAs. Images were acquired at 1 frame/h from 60-120 h.p.i. Scale bar = 10 µm. Time Stamp is in hours. Nuclei are manually traced to enhance visualization. Data shown is representative of 3 independent biological replicates.
Video 2
Tubulin acetylation is required for efficient nuclear rotation by HCMV. Expression of a non-acteylatable K40R tubulin mutant suppresses nuclear rotation. NHDFs expressing wild-type or K40R forms of tubulin were infected with TB40/E-UL99-eGFP. Images were acquired at 1 frame/h from 60-120 h.p.i. Scale bar = 10 µm. Time Stamp is in hours. Nuclei are manually traced to enhance visualization. Data shown is representative of 3 independent biological replicates.
Video 3
Mask-RCNN analysis of SUN1 polarity in HCMV-infected cells. A Mask-RCNN network can be used to classify and segment HCMV infected cells in whole-coverslip scanning datasets of HCMV infection at 96 h.p.i. (used in Figure 1g). High confidence HCMV infected cells are identified (top left), then the center of the nucleus and AC are determined from the segmentation masks (red and blue circles respectively) to instantaneously draw a 150 pixel wide linescan (white shaded line, intensity values displayed top right). This analysis is highly spatially aware and can be used to identify the intersection points between this linescan and the nucleus (white circles) which can be used to rotate and align individual nuclei (middle panels) to reveal a consistent polarization of SUN1 across a growing population of single cells using a growing average projection. Scale bar = 10 µm, n=481 cell subset is shown for brevity. Data shown is representative of 3 independent biological replicates.
Video 4
SUN1 depletion suppresses nuclear rotation by HCMV. NHDFs were infected with TB40/E-UL99-eGFP followed by treatment with the indicated siRNAs. Images were acquired at 1 frame/h from 60-120 h.p.i. Scale bar = 10 µm. Time Stamp is in hours. Nuclei are manually traced to enhance visualization. Data shown is representative of 3 independent biological replicates.
Video 5
Effects of GFP-Nesprin-2G constructs on nuclear rotation in HCMV-infected cells. NHDFs expressing the indicated GFP-Nesprin constructs were infected with TB40/E-UL99-mCherry. Images were acquired at 1 frame/h from 60-120 h.p.i. Scale bar = 10 µm. Time Stamp is in hours. Examples of rotation events above 1800 observed in AD or LEAA constructs are shown. Note that while the degree of rotation above 1800 varies, the frequency of rotation events exceeding 1800 is higher in LEAA-expressing NHDFs (Main Figure 2f-g). Data shown is representative of 5 independent biological replicates.
Video 6
Effects of BICD2 depletion on nuclear rotation by HCMV. NHDFs were infected with TB40/E-UL99-eGFP followed by treatment with the indicated siRNAs. Images were acquired at 1 frame/h from 60-120 h.p.i. Scale bar = 10 µm. Time Stamp is in hours. Nuclei are manually traced to enhance visualization. Data shown is representative of 3 independent biological replicates.
Video 7
Formation of nuclear F-actin early in HCMV infection. NHDFs expressing nuclear actin chromobody (nAC-TagGFP2) were mock infected or infected with TB40/E. Images were acquired at 1 frame/h from 5-45 h.p.i. Scale bar = 10 µm. Time Stamp is in hours. Note how actin filaments initially seed and form thick bundles that are subsequently organized into more elaborate filamentous networks as the nucleus rotates. Data shown is representative of 3 independent biological replicates.
Video 8
Reorganization of nuclear F-actin during the nuclear rotation phase of HCMV infection. NHDFs expressing nuclear actin chromobody (nAC-TagGFP2) were infected with TB40/E-UL99-mCherry. Images were acquired at 1 frame/h from 15-97 h.p.i. Scale bar = 10 µm. Time Stamp is in hours. Note that actin filaments form as nuclear rotation begins, are organized into filamentous networks during nuclear rotation, and dissipate as rotation ends. Data shown is representative of 3 independent biological replicates.
Video 9
Nuclear F-actin fomration in NHDFs in response to DNA damage. NHDFs expressing nuclear actin chromobody (nAC-TagGFP2) were treated with 50 μM Etoposide. Images were acquired at 1 frame/h from 6-48 h, with Etoposide treatment maintained during imaging. Scale bar = 10 µm. Time lapse imaging reveals that DNA damage induces the formation of thick F-actin but that filaments are not extensively reorganized. In addition, while nuclei are jostled and move, they do not significantly rotate compared with those in HCMV-infected cells. Time Stamp is in hours. Data shown is representative of 3 independent biological replicates.
Video 10
Histones are not globally polarized as nuclei rotate during HCMV infection. NHDFs expressing histone chromobody (eGFP-Histone-Nb) were infected with TB40/E UL99-mCherry. Imaged were acquired at 3 frames/h from 55.7-107.3 h.p.i. Scale bar = 10 µm. Time lapse imaging shows that the total pool of histones is dynamic but does not polarize during the nuclear rotation phase of infection by HCMV. Time Stamp is in hours. Scale bar = 10 µm. Data shown is representative of 3 independent biological replicates.
Rights and permissions
About this article
Cite this article
Procter, D.J., Furey, C., Garza-Gongora, A.G. et al. Cytoplasmic control of intranuclear polarity by human cytomegalovirus. Nature 587, 109–114 (2020). https://doi.org/10.1038/s41586-020-2714-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2714-x
This article is cited by
-
The poxvirus F17 protein counteracts mitochondrially orchestrated antiviral responses
Nature Communications (2023)
-
PIP4K2B is mechanoresponsive and controls heterochromatin-driven nuclear softening through UHRF1
Nature Communications (2023)
-
Mechanical regulation of chromatin and transcription
Nature Reviews Genetics (2022)
-
Controlling the nucleus from a distance
Nature Reviews Microbiology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.