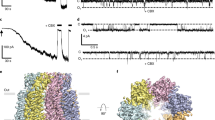
Abstract
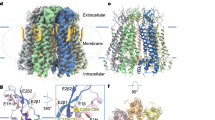
Pannexin 1 (PANX1) is an ATP-permeable channel with critical roles in a variety of physiological functions such as blood pressure regulation1, apoptotic cell clearance2 and human oocyte development3. Here we present several structures of human PANX1 in a heptameric assembly at resolutions of up to 2.8 angström, including an apo state, a caspase-7-cleaved state and a carbenoxolone-bound state. We reveal a gating mechanism that involves two ion-conducting pathways. Under normal cellular conditions, the intracellular entry of the wide main pore is physically plugged by the C-terminal tail. Small anions are conducted through narrow tunnels in the intracellular domain. These tunnels connect to the main pore and are gated by a long linker between the N-terminal helix and the first transmembrane helix. During apoptosis, the C-terminal tail is cleaved by caspase, allowing the release of ATP through the main pore. We identified a carbenoxolone-binding site embraced by W74 in the extracellular entrance and a role for carbenoxolone as a channel blocker. We identified a gap-junction-like structure using a glycosylation-deficient mutant, N255A. Our studies provide a solid foundation for understanding the molecular mechanisms underlying the channel gating and inhibition of PANX1 and related large-pore channels.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-21588 (PANX1(WT)), EMD-21589 (PANX1(ΔCTT)), EMD-21590 (CBX–PANX(ΔCTT))00, EMD-21591 (PANX1(ΔNTH/ΔCTT)), EMD-21592 (CBX–PANX1(ΔNTH/ΔCTT)), EMD-21593 (PANX1(N255A)Hemi), EMD-21594 (PANX1(N255A)Gap), EMD-21595 (apo PANX1), EMD-21596 (Ca2+–PANX1), EMD-21597 (K+–PANX1) and EMD-21598 (SMA–PANX1). Structure models have been deposited in the RCSB Protein Data Bank under accession codes 6WBF (PANX1(WT)), 6WBG (PANX1(ΔCTT)), 6WBI (CBX–PANX(ΔCTT)), 6WBK (PANX1(ΔNTH/ΔCTT)), 6WBL (CBX–PANX1(ΔNTH/ΔCTT)), 6WBM (PANX1(N255A)Hemi) and 6WBN (PANX1(N255A)Gap).
References
Billaud, M. et al. A molecular signature in the pannexin1 intracellular loop confers channel activation by the α1 adrenoreceptor in smooth muscle cells. Sci. Signal. 8, ra17 (2015).
Chekeni, F. B. et al. Pannexin 1 channels mediate ‘find-me’ signal release and membrane permeability during apoptosis. Nature 467, 863–867 (2010).
Sang, Q. et al. A pannexin 1 channelopathy causes human oocyte death. Sci. Transl. Med. 11, eaav8731 (2019).
Tozaki-Saitoh, H., Tsuda, M. & Inoue, K. Role of purinergic receptors in CNS function and neuroprotection. Adv. Pharmacol. 61, 495–528 (2011).
Ren, J. & Bertrand, P. P. Purinergic receptors and synaptic transmission in enteric neurons. Purinergic Signal. 4, 255–266 (2008).
Tsuda, M., Tozaki-Saitoh, H. & Inoue, K. Pain and purinergic signaling. Brain Res. Rev. 63, 222–232 (2010).
Le, T.-T. T. et al. Purinergic signaling in pulmonary inflammation. Front. Immunol. 10, 1633 (2019).
Zhang, Z. et al. Regulated ATP release from astrocytes through lysosome exocytosis. Nat. Cell Biol. 9, 945–953 (2007).
Dahl, G. ATP release through pannexon channels. Phil. Trans. R. Soc. Lond. B 370, 20140191 (2015).
Taruno, A. et al. CALHM1 ion channel mediates purinergic neurotransmission of sweet, bitter and umami tastes. Nature 495, 223–226 (2013).
Anselmi, F. et al. ATP release through connexin hemichannels and gap junction transfer of second messengers propagate Ca2+ signals across the inner ear. Proc. Natl Acad. Sci. USA 105, 18770–18775 (2008).
Penuela, S., Gehi, R. & Laird, D. W. The biochemistry and function of pannexin channels. Biochim. Biophys. Acta 1828, 15–22 (2013).
Adamson, S. E. et al. Pannexin 1 is required for full activation of insulin-stimulated glucose uptake in adipocytes. Mol. Metab. 4, 610–618 (2015).
Makarenkova, H. P., Shah, S. B. & Shestopalov, V. I. The two faces of pannexins: new roles in inflammation and repair. J. Inflamm. Res. 11, 273–288 (2018).
Thompson, R. J. et al. Activation of pannexin-1 hemichannels augments aberrant bursting in the hippocampus. Science 322, 1555–1559 (2008).
Silverman, W. R. et al. The pannexin 1 channel activates the inflammasome in neurons and astrocytes. J. Biol. Chem. 284, 18143–18151 (2009).
Crespo Yanguas, S. et al. Pannexin1 as mediator of inflammation and cell death. Biochim. Biophys. Acta 1864, 51–61 (2017).
Michalski, K. & Kawate, T. Carbenoxolone inhibits pannexin1 channels through interactions in the first extracellular loop. J. Gen. Physiol. 147, 165–174 (2016).
Poon, I. K. H. et al. Unexpected link between an antibiotic, pannexin channels and apoptosis. Nature 507, 329–334 (2014).
Myers, J. B. et al. Structure of native lens connexin 46/50 intercellular channels by cryo-EM. Nature 564, 372–377 (2018).
Maeda, S. et al. Structure of the connexin 26 gap junction channel at 3.5 Å resolution. Nature 458, 597–602 (2009).
Oshima, A., Tani, K. & Fujiyoshi, Y. Atomic structure of the innexin-6 gap junction channel determined by cryo-EM. Nat. Commun. 7, 13681 (2016).
Kefauver, J. M. et al. Structure of the human volume regulated anion channel. eLife 7, e38461 (2018).
Kasuya, G. et al. Cryo-EM structures of the human volume-regulated anion channel LRRC8. Nat. Struct. Mol. Biol. 25, 797–804 (2018).
Deneka, D., Sawicka, M., Lam, A. K. M., Paulino, C. & Dutzler, R. Structure of a volume-regulated anion channel of the LRRC8 family. Nature 558, 254–259 (2018).
Kern, D. M., Oh, S., Hite, R. K. & Brohawn, S. G. Cryo-EM structures of the DCPIB-inhibited volume-regulated anion channel LRRC8A in lipid nanodiscs. eLife 8, e42636 (2019).
Choi, W., Clemente, N., Sun, W., Du, J. & Lü, W. The structures and gating mechanism of human calcium homeostasis modulator 2. Nature 576, 163–167 (2019).
Syrjanen, J. L. et al. Structure and assembly of calcium homeostasis modulator proteins. Nat. Struct. Mol. Biol. 27, 150–159 (2020).
Sosinsky, G. E. et al. Pannexin channels are not gap junction hemichannels. Channels (Austin) 5, 193–197 (2011).
Boassa, D. et al. Pannexin1 channels contain a glycosylation site that targets the hexamer to the plasma membrane. J. Biol. Chem. 282, 31733–31743 (2007).
Beckmann, A., Grissmer, A., Krause, E., Tschernig, T. & Meier, C. Pannexin-1 channels show distinct morphology and no gap junction characteristics in mammalian cells. Cell Tissue Res. 363, 751–763 (2016).
Sahu, G., Sukumaran, S. & Bera, A. K. Pannexins form gap junctions with electrophysiological and pharmacological properties distinct from connexins. Sci. Rep. 4, 4955 (2014).
Sandilos, J. K. et al. Pannexin 1, an ATP release channel, is activated by caspase cleavage of its pore-associated C-terminal autoinhibitory region. J. Biol. Chem. 287, 11303–11311 (2012).
Chiu, Y.-H. et al. A quantized mechanism for activation of pannexin channels. Nat. Commun. 8, 14324 (2017).
Michalski, K., Henze, E., Nguyen, P., Lynch, P. & Kawate, T. The weak voltage dependence of pannexin 1 channels can be tuned by N-terminal modifications. J. Gen. Physiol. 150, 1758–1768 (2018).
Wang, J. et al. The membrane protein pannexin1 forms two open-channel conformations depending on the mode of activation. Sci. Signal. 7, ra69 (2014).
Locovei, S., Wang, J. & Dahl, G. Activation of pannexin 1 channels by ATP through P2Y receptors and by cytoplasmic calcium. FEBS Lett. 580, 239–244 (2006).
DeLalio, L. J. et al. Constitutive SRC-mediated phosphorylation of pannexin 1 at tyrosine 198 occurs at the plasma membrane. J. Biol. Chem. 294, 6940–6956 (2019).
Weilinger, N. L. et al. Metabotropic NMDA receptor signaling couples Src family kinases to pannexin-1 during excitotoxicity. Nat. Neurosci. 19, 432–442 (2016).
Furlow, P. W. et al. Mechanosensitive pannexin-1 channels mediate microvascular metastatic cell survival. Nat. Cell Biol. 17, 943–952 (2015).
Penuela, S., Celetti, S. J., Bhalla, R., Shao, Q. & Laird, D. W. Diverse subcellular distribution profiles of pannexin 1 and pannexin 3. Cell Commun. Adhes. 15, 133–142 (2008).
Michalski, K. et al. The cryo-EM structure of pannexin 1 reveals unique motifs for ion selection and inhibition. eLife 9, e54670 (2020).
Deng, Z. et al. Cryo-EM structures of the ATP release channel pannexin 1. Preprint at https://www.biorxiv.org/content/10.1101/2020.01.05.895235v1 (2020).
Ma, W. et al. Pannexin 1 forms an anion-selective channel. Pflugers Arch. 463, 585–592 (2012).
Wang, J. & Dahl, G. SCAM analysis of Panx1 suggests a peculiar pore structure. J. Gen. Physiol. 136, 515–527 (2010).
Romanov, R. A. et al. The ATP permeability of pannexin 1 channels in a heterologous system and in mammalian taste cells is dispensable. J. Cell Sci. 125, 5514–5523 (2012).
Goehring, A. et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc. 9, 2574–2585 (2014).
Zhou, Q. et al. Target protease specificity of the viral serpin CrmA. Analysis of five caspases. J. Biol. Chem. 272, 7797–7800 (1997).
Denault, J.-B. & Salvesen, G. S. Expression, purification, and characterization of caspases. Curr. Protoc. Protein Sci. 30, 21.13.1– 21.13.15 (2002).
Haley, E. et al. Expression and purification of the human lipid-sensitive cation channel TRPC3 for structural determination by single-particle cryo-electron microscopy. J. Vis. Exp. 143, e58754 (2019).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Zivanov, J., Nakane, T. & Scheres, S. H. W. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019).
Zivanov, J., Nakane, T. & Scheres, S. H. W. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7, 253–267 (2020).
Seminario-Vidal, L. et al. Thrombin promotes release of ATP from lung epithelial cells through coordinated activation of Rho- and Ca2+-dependent signaling pathways. J. Biol. Chem. 284, 20638–20648 (2009).
Chiu, Y.-H., Schappe, M. S., Desai, B. N. & Bayliss, D. A. Revisiting multimodal activation and channel properties of pannexin 1. J. Gen. Physiol. 150, 19–39 (2018).
Nielsen, B. S. et al. Pannexin 1 activation and inhibition is permeant-selective. J. Physiol. 598, 361–379 (2020).
Dourado, M., Wong, E. & Hackos, D. H. Pannexin-1 is blocked by its C-terminus through a delocalized non-specific interaction surface. PLoS ONE 9, e99596 (2014).
Penuela, S., Bhalla, R., Nag, K. & Laird, D. W. Glycosylation regulates pannexin intermixing and cellular localization. Mol. Biol. Cell 20, 4313–4323 (2009).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Drozdetskiy, A., Cole, C., Procter, J. & Barton, G. J. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 43, W389–W394 (2015).
Afonine, P. V. et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D 74, 814–840 (2018).
Trabuco, L. G., Villa, E., Schreiner, E., Harrison, C. B. & Schulten, K. Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods 49, 174–180 (2009).
Williams, C. J. et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
The PyMOL Molecular Graphics System v.2.1. (Schrödinger, 2020).
Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA 98, 10037–10041 (2001).
Chovancova, E. et al. CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLOS Comput. Biol. 8, e1002708 (2012).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Goddard, T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Shen, M. R. et al. Differential expression of volume-regulated anion channels during cell cycle progression of human cervical cancer cells. J. Physiol. 529, 385–394 (2000).
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
Lomize, M. A., Pogozheva, I. D., Joo, H., Mosberg, H. I. & Lomize, A. L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 (2012).
Huang, J. et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017).
Hess, B. P-LINCS: a parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 4, 116–122 (2008).
Abraham, M. J. et al. Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015).
Acknowledgements
We thank B. Roth for the initial construct screening; G. Zhao and X. Meng for the support with data collection at the David Van Andel Advanced Cryo-Electron Microscopy Suite; the HPC team of VARI for computational support; and D. Nadziejka for technical editing. W.L. is supported by the National Institutes of Health (NIH) (grant R56HL144929). J.D. is supported by a McKnight Scholar Award, a Klingenstein-Simon Scholar Award, a Sloan Research Fellowship in neuroscience and the NIH (grant R01NS111031). Z.R. is supported by an American Heart Association postdoctoral fellowship (grant 20POST35120556).
Author information
Authors and Affiliations
Contributions
W.L. and J.D. initiated and supervised the project. Z.R. performed mutagenesis, purified PANX1, prepared and screened cryo-EM samples and performed cryo-EM data collection and processing and computational simulation. I.J.O. performed electrophysiological experiments. All authors contributed in manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Stephen Brohawn and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Purification and biochemical analysis of PANX1.
a, SEC profile of PANX1 purification using GDN. b, SDS–PAGE of purified PANX1–GFP. For gel source data, see Supplementary Fig. 1a. c, FSEC experiment for caspase 3/7 cleavage of PANX1–GFP. GFP absorbance (480 nm) is shown on the y-axis. d, Caspase 7 cleavage of purified PANX1–GFP. Cleavage of CTT results in a peak shift. Tryptophan absorbance (280 nm) is shown on the y-axis. e, SMA solubilization screening of PANX1–GFP using FSEC. Three SMA polymers (SMA25010, SMA30010 and SMA40001) were tested. GFP absorbance (480 nm) is shown on the y-axis. f, SEC profile of PANX1 purification using SMA30010. Tryptophan absorbance (280 nm) is shown on the y-axis. g, Deglycosylation test of PANX1–GFP and PANX1(N255A)–GFP using PNGase F. Bands corresponding to the glycosylated and non-glycosylated PANX1 are indicated. See Supplementary Fig. 1b for gel source data. h, FSEC analysis of the PANX1 mutations used for electrophysiology recordings. Cells expressing PANX1(WT) or mutants were solubilized using GDN. Gain-of-function mutations with lower expression level are labelled. The R75E mutant contains a peak position shifted to the right. i, j, FSEC analysis on extracellular-gate mutations of PANX1 solubilized using GDN (i) or SMA30010 (j). Peak positions of correctly assembled PANX1 and incorrectly assembled PANX1 are indicated by arrows and vertical bars. The W74R, R75D and R75E mutants showed decreased stability relative to wild type because, when extracted using detergent, they mostly ran at positions representing incorrect assemblies (i). Nevertheless, the SMA-extracted W74R, R75D and R75E mutants still showed peaks at positions representing correct assemblies (j), indicating that they are able to form correctly assembled channel complex in a native lipid environment.
Extended Data Fig. 2 The workflow of cryo-EM data processing of PANX1 constructs that do not form gap junctions.
a, The data-analysis pipeline for PANX1 cryo-EM analysis with no gap junction. Special attention was paid to determine whether the dataset indeed adheres to C7 symmetry. Two examples of PANX1 adopting a symmetric conformation (PANX1(ΔCTT)) or a non-symmetric conformation (CBX–PANX1(ΔCTT)) are shown. A detailed description of the data-analysis procedure can be found in Methods. b, The overlay of the C1 refined maps of PANX1(ΔCTT), CBX–PANX1(ΔCTT), PANX1(ΔNTH/ΔCTT) and CBX–PANX1(ΔNTH/ΔCTT) with the symmetric model of PANX1. The CBX–PANX1(ΔCTT), PANX1((ΔNTH/ΔCTT)) and CBX–PANX1(ΔNTH/ΔCTT) maps adopt a non-symmetric shape.
Extended Data Fig. 3 Representative micrographs, 2D class averages and Fourier shell correlation curves for all datasets in this study.
a–j, For each dataset, a representative micrograph, four 2D class averages and Fourier shell correlation (FSC) curve plot are shown, except for PANX1(N255A) dataset, in which two structures are shown. The map resolution is determined on the basis of the gold-standard 0.143 criterion. If an atomic model is available for the dataset, a model versus map FSC curve is also provided. The model versus map resolution is determined on the basis of the 0.5 FSC criterion. In a, a slice view of the SMA–PANX1 map showing the organization of the TMD is shown.
Extended Data Fig. 4 Local resolution estimation and representative densities.
a, b, PANX1 map. c, d, The PANX1(N255A)Gap map. To provide better visualization on the exterior and interior map quality, both non-sliced and sliced views of the maps are shown. The colour represents the local resolution in Å. Representative densities are shown for a few selected secondary structure elements of PANX1.
Extended Data Fig. 5 The workflow of cryo-EM data processing for PANX1(N255A).
To separate gap junction particles from hemichannel particles, we relied on 2D classification to distinguish tilted and side views. The top and down views were separated during 3D classification. A more detailed description of the data-analysis pipeline can be found in the Methods.
Extended Data Fig. 6 The structures of PANX1(ΔNTH/ΔCTT).
a, The apo state. b, PANX1(ΔNTH/ΔCTT) in complex with CBX. CBX is shown in orange. In a, b, odd- and even-numbered subunits are shown in blue and white, respectively; the seventh subunit is in green. Cryo-EM maps viewed parallel to the membrane (left) and from the extracellular side (middle). The unsharpened map is shown as a transparent envelope. Right, structural models viewed from the intracellular side. c, The slice view of the extracellular entrance of PANX1(ΔNTH/ΔCTT) (left) and CBX–PANX1(ΔNTH/ΔCTT) (right) maps. The CBX molecule and the side chain of W74 are shown in stick.
Extended Data Fig. 7 Patch-clamp analysis of the wild-type PANX1 and its mutants.
a, Representative traces of whole-cell current density from patch-clamped tsA201 control cells (n = 6) and tsA201 cells overexpressing: wild type (n = 12) and ΔNTH (n = 5), W74A (n = 8), R75E (n = 5), N255A (n = 5), Δ21–23 (n = 6), Δ21–27 (n = 7), R29A (n = 9), A33W (n = 10), A33W/ΔCTT (n = 4), ΔCTT (n = 4) and A33C (n = 5) mutant PANX1. Voltage steps (0.25 s) of 20 mV were imposed from −100 mV to +80 mV from a holding potential of −10 mV. Cells were first measured in standard bath solution and then re-measured following the superfusion of a bath solution containing 0.1 mM carbenoxolone. b, Mean current measured at 5 ms of experiments in a plotted as a function of clamp voltage. c, Current amplitudes of experiments in a with and without CBX. Each paired point represents an individual cell and the bar represents the mean. d, Plot of zero-current reversal potentials using a 2-s voltage ramp for various bath solutions containing either (in mM): 145 NaCl, 145 NaI, 145 sodium gluconate, 14.5 NaCl or 145 NMDG-Cl (see Methods for complete solutions). The number of cells patched (indicated in parenthesis) for the various bath solutions were as follows, 145 mM NaCl: wild type (5), R29A (7), W74A (5), R75E (7); 145 mM NaI: wild type (5), R29A (7), W74A (5), R75E (7); 145 mM sodium gluconate: wild type (5), R29A (7), W74A (5), R75E (7); 14.5 mM NaCl: wild type (4), R29A (4), W74A (5), R75E (5); 145 mM NMDG-Cl: wild type (3), R29A (2), W74A (5), R75E (3). e, f, Plots of the calculated (Methods) permeability of iodide relative to chloride (PI/PCl), the permeability of gluconate relative to chloride (Pgluconate/PCl) and permeability of sodium relative to chloride (PNa/PCl) for wild-type and mutant channels. For statistical comparisons to wild type, one-way analysis of variance with Bonferroni correction was performed. For PI/PCl, P = 0.99 (R29A), 3.7 × 10−5 (W74A) and 5.9 × 10−5 (R75E). For Pgluconate/PCl, P = 0.046 (R29A), 0.99 (W74A) and 6.8 × 10−9 (R75E). For PNa/PCl, P = 0.99 (R29A), 0.99 (W74A) and 2.2 × 10−6 (R75E). Each point represents an individual cell. The bar represents mean and error bars show s.e.m.
Extended Data Fig. 8 Comparison of large-pore channels.
a, The structures of large-pore channels, viewed parallel (top) or perpendicular (bottom) to the membrane. One subunit (or one pair of subunits) is in green. The diameter of VRAC is calculated without the cytoplasmic leucine-rich repeat domain. b, Organization of the TMD, viewed from the intracellular side. The NTH and transmembrane helices S1, S2, S3 and S4 are labelled for two subunits. Only CALHM2 has its transmembrane helices arranged in a clockwise manner. The contact between adjacent TMDs in PANX1 is made by the NTH with the S1 and S2 helices in the neighbouring subunit; the same contact in CALHM2 is made by the S2 and S4 of adjacent subunits, and in connexin by the S1 and S2 of adjacent subunits. There is no major contact in innexin and VRAC.
Extended Data Fig. 9 Secondary structure arrangement and sequence alignment.
Secondary structures based on the PANX1 structure model are labelled. The W74 forming the extracellular entrance is marked with an arrow. Key residues forming the side tunnel are labelled with a red asterisk. The cysteine residues forming the extracellular disulfide bonds are highlighted by an orange dot. The N255 glycosylation site is marked with a green dot. The gap junction interface and caspase 3/7 cleavage site are indicated with a red frame. A gain-of-function disease mutation (Δ21–23) of PANX1 is also marked.
Supplementary information
Supplementary Figure 1
This file contains the raw gel images for Extended Data Fig. 1b and 1g.
Rights and permissions
About this article
Cite this article
Ruan, Z., Orozco, I.J., Du, J. et al. Structures of human pannexin 1 reveal ion pathways and mechanism of gating. Nature 584, 646–651 (2020). https://doi.org/10.1038/s41586-020-2357-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2357-y
This article is cited by
-
Cryo-EM structures of pannexin 1 and 3 reveal differences among pannexin isoforms
Nature Communications (2024)
-
Targeting Pannexin-1 Channels: Addressing the ‘Gap’ in Chronic Pain
CNS Drugs (2024)
-
The small molecule raptinal can simultaneously induce apoptosis and inhibit PANX1 activity
Cell Death & Disease (2024)
-
Structure of a volume-regulated heteromeric LRRC8A/C channel
Nature Structural & Molecular Biology (2023)
-
Cryo-EM structure of human heptameric pannexin 2 channel
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.