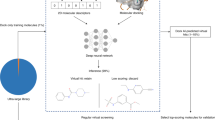
Abstract
On average, an approved drug currently costs US$2–3 billion and takes more than 10 years to develop1. In part, this is due to expensive and time-consuming wet-laboratory experiments, poor initial hit compounds and the high attrition rates in the (pre-)clinical phases. Structure-based virtual screening has the potential to mitigate these problems. With structure-based virtual screening, the quality of the hits improves with the number of compounds screened2. However, despite the fact that large databases of compounds exist, the ability to carry out large-scale structure-based virtual screening on computer clusters in an accessible, efficient and flexible manner has remained difficult. Here we describe VirtualFlow, a highly automated and versatile open-source platform with perfect scaling behaviour that is able to prepare and efficiently screen ultra-large libraries of compounds. VirtualFlow is able to use a variety of the most powerful docking programs. Using VirtualFlow, we prepared one of the largest and freely available ready-to-dock ligand libraries, with more than 1.4 billion commercially available molecules. To demonstrate the power of VirtualFlow, we screened more than 1 billion compounds and identified a set of structurally diverse molecules that bind to KEAP1 with submicromolar affinity. One of the lead inhibitors (iKeap1) engages KEAP1 with nanomolar affinity (dissociation constant (Kd) = 114 nM) and disrupts the interaction between KEAP1 and the transcription factor NRF2. This illustrates the potential of VirtualFlow to access vast regions of the chemical space and identify molecules that bind with high affinity to target proteins.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The ready-to-dock library from Enamine is freely available online on the homepage of VirtualFlow at http://virtual-flow.org/real-library. Source Data for Figs. 2, 3 and Extended Data Figs. 7, 8 are available with the paper.
Code availability
VirtualFlow is mainly written in Bash (a Turing complete command language), which not only makes it simple for anyone to modify and extend the code, but also has essentially no computational overhead and is readily available in any major Linux distribution. The code for VirtualFlow is freely available on https://github.com/VirtualFlow, distributed under the GNU GPL open-source licence. The primary homepage for end users, which includes additional resources such as documentation, ligand libraries, tutorials and video demonstrations, is available at https://www.virtual-flow.org. The external docking programs discussed here are available as follows: AutoDock Vina is available at http://vina.scripps.edu, QuickVina 2 and QuickVina-W at https://qvina.github.io, Vina-Carb at http://glycam.org/docs/othertoolsservice/download-docs/publication-materials/vina-carb, Smina at https://sourceforge.net/projects/smina, AutoDockFR at http://adfr.scripps.edu and VinaXB at https://github.com/ssirimulla/vinaXB.
References
DiMasi, J. A., Grabowski, H. G. & Hansen, R. W. Innovation in the pharmaceutical industry: new estimates of R&D costs. J. Health Econ. 47, 20–33 (2016).
Lyu, J. et al. Ultra-large library docking for discovering new chemotypes. Nature 566, 224–229 (2019).
Zhang, S., Kumar, K., Jiang, X., Wallqvist, A. & Reifman, J. DOVIS: an implementation for high-throughput virtual screening using AutoDock. BMC Bioinformatics 9, 126 (2008).
Jiang, X., Kumar, K., Hu, X., Wallqvist, A. & Reifman, J. DOVIS 2.0: an efficient and easy to use parallel virtual screening tool based on AutoDock 4.0. Chem. Cent. J. 2, 18 (2008).
Hassan, N. M., Alhossary, A. A., Mu, Y. & Kwoh, C.-K. Protein-ligand blind docking using QuickVina-W with inter-process spatio-temporal integration. Sci. Rep. 7, 15451 (2017).
Bohacek, R. S., McMartin, C. & Guida, W. C. The art and practice of structure-based drug design: a molecular modeling perspective. Med. Res. Rev. 16, 3–50 (1996).
Yonchuk, J. G. et al. Characterization of the potent, selective Nrf2 activator, 3-(pyridin-3-ylsulfonyl)-5-(trifluoromethyl)-2H-chromen-2-one, in cellular and in vivo models of pulmonary oxidative stress. J. Pharmacol. Exp. Ther. 363, 114–125 (2017).
Pallesen, J. S., Tran, K. T. & Bach, A. Non-covalent small-molecule Kelch-like ECH-associated protein 1-nuclear factor erythroid 2-related factor 2 (Keap1–Nrf2) inhibitors and their potential for targeting central nervous system diseases. J. Med. Chem. 61, 8088–8103 (2018).
Davies, T. G. et al. Monoacidic inhibitors of the Kelch-like ECH-associated protein 1: nuclear factor erythroid 2-related factor 2 (KEAP1:NRF2) protein–protein interaction with high cell potency identified by fragment-based discovery. J. Med. Chem. 59, 3991–4006 (2016).
Cuadrado, A. et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 18, 295–317 (2019).
Sterling, T. & Irwin, J. J. ZINC 15—ligand discovery for everyone. J. Chem. Inf. Model. 55, 2324–2337 (2015).
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Alhossary, A., Handoko, S. D., Mu, Y. & Kwoh, C.-K. Fast, accurate, and reliable molecular docking with QuickVina 2. Bioinformatics 31, 2214–2216 (2015).
Koes, D. R., Baumgartner, M. P. & Camacho, C. J. Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. J. Chem. Inf. Model. 53, 1893–1904 (2013).
Ravindranath, P. A., Forli, S., Goodsell, D. S., Olson, A. J. & Sanner, M. F. AutoDockFR: advances in protein-ligand docking with explicitly specified binding site flexibility. PLOS Comput. Biol. 11, e1004586 (2015).
Koebel, M. R., Schmadeke, G., Posner, R. G. & Sirimulla, S. AutoDock VinaXB: implementation of XBSF, new empirical halogen bond scoring function, into AutoDock Vina. J. Cheminform. 8, 27 (2016).
Nivedha, A. K., Thieker, D. F., Makeneni, S., Hu, H. & Woods, R. J. Vina-Carb: improving glycosidic angles during carbohydrate docking. J. Chem. Theory Comput. 12, 892–901 (2016).
Amaro, R. E. et al. Ensemble docking in drug discovery. Biophys. J. 114, 2271–2278 (2018).
Houston, D. R. & Walkinshaw, M. D. Consensus docking: improving the reliability of docking in a virtual screening context. J. Chem. Inf. Model. 53, 384–390 (2013).
Marcotte, D. et al. Small molecules inhibit the interaction of Nrf2 and the Keap1 Kelch domain through a non-covalent mechanism. Bioorg. Med. Chem. 21, 4011–4019 (2013).
Andrei, S. A. et al. Stabilization of protein–protein interactions in drug discovery. Expert Opin. Drug Discov. 12, 925–940 (2017).
Ragoza, M., Hochuli, J., Idrobo, E., Sunseri, J. & Koes, D. R. Protein-ligand scoring with convolutional neural networks. J. Chem. Inf. Model. 57, 942–957 (2017).
Reymond, J. L. The chemical space project. Acc. Chem. Res. 48, 722–730 (2015).
O’Boyle, N. M. et al. Open Babel: an open chemical toolbox. J. Cheminform. 3, 33 (2011).
Morris, G. M. et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 (2009).
Hutsell, S. Q., Kimple, R. J., Siderovski, D. P., Willard, F.S. & Kimple, A. J. High-affinity immobilization of proteins using biotin- and GST-based coupling strategies. Methods Mol. Biol. 627, 75–90 (2010).
Hämäläinen, M. D. et al. Label-free primary screening and affinity ranking of fragment libraries using parallel analysis of protein panels. J. Biomol. Screen. 13, 202–209 (2008).
Hulme, E. C. (ed.) Receptor–Ligand Interactions: A Practical Approach (Oxford Univ. Press, 1992).
Gans, P. et al. Stereospecific isotopic labeling of methyl groups for NMR spectroscopic studies of high-molecular-weight proteins. Angew. Chem. Int. Ed. 49, 1958–1962 (2010).
Lu, M. et al. Discovery of a Keap1-dependent peptide PROTAC to knockdown Tau by ubiquitination-proteasome degradation pathway. Eur. J. Med. Chem. 146, 251–259 (2018).
Irwin, J. J. et al. An aggregation advisor for ligand discovery. J. Med. Chem. 58, 7076–7087 (2015).
LaPlante, S. R. et al. Compound aggregation in drug discovery: implementing a practical NMR assay for medicinal chemists. J. Med. Chem. 56, 5142–5150 (2013).
Baell, J. B. & Holloway, G. A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 53, 2719–2740 (2010).
Baell, J. B. & Nissink, J. W. M. Seven year itch: pan-assay interference compounds (PAINS) in 2017—utility and limitations. ACS Chem. Biol. 13, 36–44 (2018).
Capuzzi, S. J., Muratov, E. N. & Tropsha, A. Phantom PAINS: problems with the utility of alerts for pan-assay interference compounds. J. Chem. Inf. Model. 57, 417–427 (2017).
Acknowledgements
We thank M. Zhang for help with the binding assays; the research computing teams of the Faculty of Arts and Sciences at Harvard University (especially S. Yockel, J. Cuff, F. Pontiggia and P. Edmon), the Jülich Supercomputing Centre, the Freie Universität (especially J. Dreger), the Harvard Medical School (HMS), the HLRN and the IT support of HMS (especially K. Bayer, G. Sekmokas and D. Morgan) for their support; K. E. Leigh, N. Gray, M. Kostic, A. Dubey, B. Klein, S. Schwaninger and S. Wu for discussions and manuscript preparation; the ICCB-Longwood Screening and East Quad NMR Facilities at HMS for assistance with the ligand screen; K. Arnett and the Center for Macromolecular Interactions at the HMS for advice on the SPR and BLI experiments; A. Jaffe for his support; and the teams from the Google Cloud Platform (especially S. Fang, R. Goldenbroit and D. Payne), Amazon Web Services, and Fluid Numerics for their support. This work was partially funded by a scholarship to C.G. from the Max Planck Institute for Molecular Genetics in Berlin and a scholarship from the Einstein Center for Mathematics Berlin. C.G. and K.F. thank the ECMath and MATHEON. C.G. is grateful to C. Schütte and P. Imhof for their support and supervision during his doctoral studies. We thank Z. Alirezaeizanjani, M. Bagherpoor and Anita Nivedha for testing VirtualFlow. M.H. acknowledges funding from Deutsche Forschungsgemeinschaft (CRC 958/Project A04, CRC 1114/Project A04). A.B. was supported by an Austrian Science Fund’s Schrödinger Fellowship (J3872-B21) and an American Heart Association’s fellowship (19POST34380800). This research was supported in part by grant TRT 0159 from the Templeton Religion Trust and by ARO Grant W911NF1910302 to A. Jaffe. K.M.P.D. was supported by a fellowship from the Max Kade Foundation and the Austrian Academy of Sciences. H.A. acknowledges funding from the Claudia Adams Barr Program for Innovative Cancer Research. G.W. acknowledges support from NIH grant CA200913, AI037581 and GM129026.
Author information
Authors and Affiliations
Contributions
C.G. conceived the project, and designed and implemented the drug discovery platform (VirtualFlow). H.A. and C.G. designed the experimental workflow. A.B., H.A., Z.-F.W., P.D.F. and K.M.P.D. designed and carried out the fluorescence polarization and NMR experiments. Z.-F.W. designed and carried out the SPR and BLI experiments. K.M.P.D. and Z.-F.W. carried out the dynamic light scattering experiments. M.H. provided technical assistance regarding the code and homepage. C.G. designed the applications (screening of KEAP1 and the preparation of the REAL library). P.W.C. analysed the NMR data. C.G. carried out the computations using VFLP (preparation of the REAL database) and VFVS (screening/rescoring of KEAP1). Y.S. Malets created the web interface to the REAL database. C.G. prepared the VirtualFlow homepage. Y.S. Moroz prepared the REAL database in the initial SMILES format. C.G. and Y.S. Moroz designed the structure of the VirtualFlow version of the REAL database. Y.S. Moroz, I.I. and D.S.R. supervised and directed the synthesis and purification of the on-demand compounds from the REAL library. D.A.S. helped to evaluate the screening hits. C.G., H.A., A.B., K.F., Z.-F.W., P.W.C. and G.W. prepared the manuscript. K.F., H.A. and G.W. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
I.I., D.S.R. and Y. S. Malets work for Enamine, a company that is involved in the synthesis and distribution of drug-like compounds. Y. S. Moroz is a scientific advisor for Enamine.
Additional information
Peer review information Nature thanks Tara Mirzadegan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Schematic overview of the organization of the VirtualFlow workflow on computer clusters.
A computer cluster consists of compute nodes, that is, single computers (blue boxes), which contain a certain number of CPU cores (black squares inside the blue boxes). The resource manager (batch system) of the cluster generates so-called jobs (large violet ovals), each of which uses a certain number of CPU cores and nodes. In the example, each job uses three compute nodes, in which each node has eight CPU cores. Each job can contain multiple sub-jobs, referred to as job steps (purple circles). With VirtualFlow, each job step comprises multiple queues (white oval shapes within the purple circles). Often the workflow is set up such that on each CPU core one queue is running. Hierarchical multi-organization is required to allow VirtualFlow to run on any type of cluster, from the largest supercomputers (which often require that a single job has multiple nodes) to very small clusters (which often allow a job to use single CPU cores). Each queue processes ligands, which are taken from the input collections in raw form and stored in the output collection or database. The central task list contains all of the ligand collections that should be processed by the workflow, and they are distributed among the queues (into local task lists) by a workload balancer at the beginning of each job. The user can choose any number of batch system jobs (first row comprising job 1.1 to job X.1), which will automatically start successive jobs (second row comprising job 1.2 to job X.2) after their completion.
Extended Data Fig. 2 Overview of possible processing steps during ligand preparation with VFLP.
Ligands can be desalted, neutralized, and one, or possibly multiple, tautomeric state(s) as well as protonation states for each tautomer computed at specific pH values can be generated, three-dimensional coordinates can be computed and, finally, the molecules can be converted into one or potentially multiple desired target formats.
Extended Data Fig. 3 Docking and virtual screening metrics.
a, Scaling behaviour of VFVS using QuickVina 2 as the docking program. Tests with up to 30,000 cores on two local computer clusters (LC1 and LC2) and up to 160,000 CPUs on the GCP were carried out. The measured speedup is linear. DOVIS 2.0, an alternative software for virtual screenings on Linux computer clusters using AutoDock, was shown to exhibit near-linear scaling only up to 256 cores, as previously reported4. b, The computational time required (in days) for VFVS to complete virtual screens of different sizes, as a function of the number of CPUs being used in parallel. Each curve corresponds to a input ligand library with a different size, and the average computation time per ligand was assumed to be 5 s per ligand. c, Docking time of an average-sized ligand on a modern Intel CPU (using only a single core) as a function of the exhaustiveness parameter for different docking programs supported by VFVS. The bar plot in the inset shows the slope of the curves, which corresponds to the docking time per exhaustiveness unit. The test ligand that was used for this purpose is given by the SMILES code CN1CCN(S(=O)(=O)N2CCN(C(=O)CCCNC(=O)C3CC3)CC2)CC1. More detailed benchmarks can be found in publications related to these docking programs5,12,13,14,15,16,17.
Extended Data Fig. 4 Binding of the NRF2 peptide to KEAP1 as assayed by fluorescence polarization and BLI.
a, a TAMRA-tagged NRF2 peptide was used for the fluorescence polarization (FP) assay. The fluorescence polarization assay was performed with three technical replicates per point. Data are mean ± s.d. for each titration point, along with the fitted curve. Two independent experiments were performed, each with similar results and one representative result is shown. b, A biotin-tagged NRF2 peptide was used for the BLI assay. The BLI experiment was repeated independently twicewith similar results and one representative result is shown.
Extended Data Fig. 5 Comparison of iKeap1 with the previously identified displacer C17.
a, Crystal structure (PDB ID: 5FNQ)9 of KEAP1 with its ligand removed, the structure used for the primary virtual screening procedure. b, Crystal structure of KEAP1 (PDB ID: 4IQK) with ligand C17 (Supplementary Table 1), the chemical structure of which is shown in d. c, d, iKeap1, the best displacer of the NFR2 peptide (c), is similar to compound C17, which has previously been identified by experimental methods (d). Although iKeap1 and C17 look similar, they differ in a number of aspects in their core scaffold (thus, analogues of the two compounds cover distinct chemical spaces, assuming that the analogues retain the core scaffold of the parent compound). This similarity, as well as the fact that the predicted docking positions (Fig. 3a) of both ligands (b) are nearly identical, is additional evidence that iKeap1 is binding at the predicted site.
Extended Data Fig. 6 Overview of binding assays to determine the activity of the hits identified by VirtualFlow.
This schematic outlines the experimental validation workflow. The binding experiments can be broadly classified into two categories: (i) assays that directly detect the binding of the compounds to KEAP1 (SPR and NMR) and (ii) assays that detect the displacement of the NRF2 peptide from KEAP1 (fluorescence polarization and BLI). Compounds in level 2 SPR experiments were classified as active if they exhibited dose-dependent activity (measured over a range of five concentrations) and had an RU value greater than 4 at a compound concentration of 20 μM. a, The high-throughput workflow in which the 590 compounds identified as hits by VirtualFlow were tested using SPR and fluorescence polarization. The hits identified here were further validated by BLI and the potential of these hits to form aggregates was tested by DLS. b, Then, 23 of the potent hits were chosen for level 3 SPR analysis to measure accurate binding affinities. c, Six of the potent binders were further subjected to NMR analysis in both protein-detected and ligand-detected NMR experiments.
Extended Data Fig. 7 Binder versus displacer.
Here we highlight two scaffolds, iKeap8 and iKeap9, to illustrate the difference between binders and displacers. a, b, SPR confirms that both iKeap8 (a) and iKeap9 (b) bind to KEAP1 and with similar Kd values. Data are representative results from the SPR assay for iKeap8 and iKeap9. For each compound, three independent SPR experiments were performed, each with similar results and one representative result is shown. c, d, Ligand-detection NMR experiments shows that both iKeap8 (c) and iKeap9 (d) bind to KEAP1. e–h, However, fluorescence polarization (e, f) and BLI (g, h) assays show that iKeap8 (e, g) is able to displace the NRF2 peptide whereas iKeap9 (f, h) is not able to effectively displace the NRF2 peptide. The fluorescence polarization assay was performed with three technical replicates per concentration measured. Data are mean ± s.d. for each titration point shown together with the fitted curve.
Extended Data Fig. 8 Displacers validated by fluorescence polarization and BLI.
Here we show two more displacers, iKeap7 and iKeap22. a, b, Both iKeap7 (a) and iKeap22 (b) were confirmed as binders by SPR. c, d, Ligand-detection NMR experiments show that both iKeap7 (c) and iKeap22 (d) bind to KEAP1. e, iKeap7 is confirmed to be a displacer of the NRF2 peptide by both fluorescence polarization and BLI (data not shown). f, As the fluorescence polarization experiments of iKeap22 were affected by autofluorescence, BLI was needed to confirm that this compounds is a displacer. The fluorescence polarization assay was performed with three technical replicates per concentration measured. Data are mean ± s.d. for each titration point, shown along with the fitted curve. Two independent BLI experiments were performed with similar results and one representative result is shown here.
Extended Data Fig. 9 NRF2 peptide- and ligand-binding sites, rationale for binder versus displacer.
Here we show the docking pose of one of the hit compounds (iKeap9, green ball-and-stick representation) bound to KEAP1, together with the NRF2 peptide (PDB ID: 4IFL; peptide in violet). iKeap9 is a tight binder (180 nM by steady-state SPR) but cannot displace NRF2. Left, the top view. Right, the side view of the cross-section of KEAP1 along the central plane. The violet box indicates the docking region (where the ligands were allowed to bind), which was used in the virtual screening. The site of interest includes a part of the deep pocket/tunnel of the β-barrel-shaped KEAP1, as it enables ligands to bind more tightly by insertion into the channel than on a shallow surface. However, the deep tunnel is largely non-overlapping with the peptide-binding site (which binds to the entrance site of the tunnel). Thus, binding molecules might only partially interfere with peptide binding, which could reduce or eliminate the ability of small-molecule binders to displace the peptide. The ability of a small molecule to displace the peptide is difficult to predict, and was not attempted in this study. In some cases, small molecules can also act as molecular glues and strengthen the interaction between NRF2 and KEAP1.
Supplementary information
Supplementary Information
Supplementary information for the main manuscript, containing additional experimental data, a probabilistic model of the true hit rate, the VirtualFlow version of the REAL library, and additional data about VirtualFlow.
Rights and permissions
About this article
Cite this article
Gorgulla, C., Boeszoermenyi, A., Wang, ZF. et al. An open-source drug discovery platform enables ultra-large virtual screens. Nature 580, 663–668 (2020). https://doi.org/10.1038/s41586-020-2117-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2117-z
This article is cited by
-
Computational drug development for membrane protein targets
Nature Biotechnology (2024)
-
Integrating QSAR modelling and deep learning in drug discovery: the emergence of deep QSAR
Nature Reviews Drug Discovery (2024)
-
Curation and cheminformatics analysis of a Ugi-reaction derived library (URDL) of synthetically tractable small molecules for virtual screening application
Molecular Diversity (2024)
-
Augmented ant colony algorithm for virtual drug discovery
Journal of Mathematical Chemistry (2024)
-
LS-HTC: an HTC system for large-scale jobs
CCF Transactions on High Performance Computing (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.