Abstract
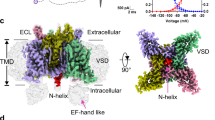
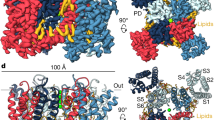
Inactivation is the process by which ion channels terminate ion flux through their pores while the opening stimulus is still present1. In neurons, inactivation of both sodium and potassium channels is crucial for the generation of action potentials and regulation of firing frequency1,2. A cytoplasmic domain of either the channel or an accessory subunit is thought to plug the open pore to inactivate the channel via a ‘ball-and-chain’ mechanism3,4,5,6,7. Here we use cryo-electron microscopy to identify the molecular gating mechanism in calcium-activated potassium channels by obtaining structures of the MthK channel from Methanobacterium thermoautotrophicum—a purely calcium-gated and inactivating channel—in a lipid environment. In the absence of Ca2+, we obtained a single structure in a closed state, which was shown by atomistic simulations to be highly flexible in lipid bilayers at ambient temperature, with large rocking motions of the gating ring and bending of pore-lining helices. In Ca2+-bound conditions, we obtained several structures, including multiple open-inactivated conformations, further indication of a highly dynamic protein. These different channel conformations are distinguished by rocking of the gating rings with respect to the transmembrane region, indicating symmetry breakage across the channel. Furthermore, in all conformations displaying open channel pores, the N terminus of one subunit of the channel tetramer sticks into the pore and plugs it, with free energy simulations showing that this is a strong interaction. Deletion of this N terminus leads to functionally non-inactivating channels and structures of open states without a pore plug, indicating that this previously unresolved N-terminal peptide is responsible for a ball-and-chain inactivation mechanism.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The map of calcium-free MthK and the four maps of calcium-bound MthK have been deposited in the Electron Microscopy Data Bank (EMDB) under accession codes EMD-20663, EMD-20662, EMD-20664, EMD-20665 and EMD-20653, respectively. The maps for the two calcium-bound RCK gating rings have been deposited with EMDB under accession codes EMD-20652 and EMD-20650, respectively. The maps for calcium-bound MthK(Δ2–17) full-length states and RCK gating-ring states have been deposited with accession codes EMD-20930, EMD-20931, EMD-20932, EMD-20925 and EMD-20929, respectively. Atomic coordinates for the calcium-free MthK, calcium-bound MthK and the two calcium-bound RCK gating rings have been deposited with the Protein Data Bank with accession codes 6U6D, 6U68, 6U6E, 6U6H, 6U5R, 6U5P and 6U5N, respectively. Atomic coordinates for calcium-bound MthK(Δ2–17) full-length states and RCK gating-ring states have been deposited with accession codes 6UX7, 6UXA, 6UXB, 6UWN and 6UX4. Extended Data Fig. 9 contains raw single-channel and stopped-flow fluorescence decay data, which are available from the corresponding author upon request.
References
Hille, B. Ion Channels of Excitable Membranes 3rd edn (Sinauer Associates, 2001).
Yellen, G. The moving parts of voltage-gated ion channels. Q. Rev. Biophys. 31, 239–295 (1998).
Hoshi, T., Zagotta, W. N. & Aldrich, R. W. Biophysical and molecular mechanisms of Shaker potassium channel inactivation. Science 250, 533–538 (1990).
Armstrong, C. M., Bezanilla, F. & Rojas, E. Destruction of sodium conductance inactivation in squid axons perfused with pronase. J. Gen. Physiol. 62, 375–391 (1973).
Bezanilla, F. & Armstrong, C. M. Inactivation of the sodium channel. I. Sodium current experiments. J. Gen. Physiol. 70, 549–566 (1977).
Wallner, M., Meera, P. & Toro, L. Molecular basis of fast inactivation in voltage and Ca2+-activated K+ channels: a transmembrane β-subunit homolog. Proc. Natl Acad. Sci. USA 96, 4137–4142 (1999).
Zhou, M., Morais-Cabral, J. H., Mann, S. & MacKinnon, R. Potassium channel receptor site for the inactivation gate and quaternary amine inhibitors. Nature 411, 657–661 (2001).
Berridge, M. J., Bootman, M. D. & Roderick, H. L. Calcium signalling: dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 4, 517–529 (2003).
Südhof, T. C. Calcium control of neurotransmitter release. Cold Spring Harb. Perspect. Biol. 4, a011353 (2012).
Yang, H., Zhang, G. & Cui, J. BK channels: multiple sensors, one activation gate. Front. Physiol. 6, 29 (2015).
Geng, Y. & Magleby, K. L. Single-channel kinetics of BK (Slo1) channels. Front. Physiol. 5, 532 (2015).
Vergara, C., Latorre, R., Marrion, N. V. & Adelman, J. P. Calcium-activated potassium channels. Curr. Opin. Neurobiol. 8, 321–329 (1998).
Hite, R. K., Tao, X. & MacKinnon, R. Structural basis for gating the high-conductance Ca2+-activated K+ channel. Nature 541, 52–57 (2017).
Tao, X., Hite, R. K. & MacKinnon, R. Cryo-EM structure of the open high-conductance Ca2+-activated K+ channel. Nature 541, 46–51 (2017).
Zhou, Y., Yang, H., Cui, J. & Lingle, C. J. Threading the biophysics of mammalian Slo1 channels onto structures of an invertebrate Slo1 channel. J. Gen. Physiol. 149, 985–1007 (2017).
Li, W. & Aldrich, R. W. State-dependent block of BK channels by synthesized Shaker ball peptides. J. Gen. Physiol. 128, 423–441 (2006).
Zhou, Y., Xia, X. M. & Lingle, C. J. Cysteine scanning and modification reveal major differences between BK channels and Kv channels in the inner pore region. Proc. Natl Acad. Sci. USA 108, 12161–12166 (2011).
Tang, Q. Y., Zeng, X. H. & Lingle, C. J. Closed-channel block of BK potassium channels by bbTBA requires partial activation. J. Gen. Physiol. 134, 409–436 (2009).
Jiang, Y. et al. The open pore conformation of potassium channels. Nature 417, 523–526 (2002).
Jiang, Y. et al. Crystal structure and mechanism of a calcium-gated potassium channel. Nature 417, 515–522 (2002).
Ye, S., Li, Y., Chen, L. & Jiang, Y. Crystal structures of a ligand-free MthK gating ring: insights into the ligand gating mechanism of K+ channels. Cell 126, 1161–1173 (2006).
Yuan, P., Leonetti, M. D., Hsiung, Y. & MacKinnon, R. Open structure of the Ca2+ gating ring in the high-conductance Ca2+-activated K+ channel. Nature 481, 94–97 (2012).
Lorenzo-Ceballos, Y., Carrasquel-Ursulaez, W., Castillo, K., Alvarez, O. & Latorre, R. Calcium-driven regulation of voltage-sensing domains in BK channels. eLife 8, e44934 (2019).
Miranda, P., Holmgren, M. & Giraldez, T. Voltage-dependent dynamics of the BK channel cytosolic gating ring are coupled to the membrane-embedded voltage sensor. eLife 7, e40664 (2018).
Zhang, G. et al. Deletion of cytosolic gating ring decreases gate and voltage sensor coupling in BK channels. J. Gen. Physiol. 149, 373–387 (2017).
Barrett, J. N., Magleby, K. L. & Pallotta, B. S. Properties of single calcium-activated potassium channels in cultured rat muscle. J. Physiol. 331, 211–230 (1982).
McManus, O. B. & Magleby, K. L. Kinetic states and modes of single large-conductance calcium-activated potassium channels in cultured rat skeletal muscle. J. Physiol. 402, 79–120 (1988).
Zadek, B. & Nimigean, C. M. Calcium-dependent gating of MthK, a prokaryotic potassium channel. J. Gen. Physiol. 127, 673–685 (2006).
Li, Y., Berke, I., Chen, L. & Jiang, Y. Gating and inward rectifying properties of the MthK K+ channel with and without the gating ring. J. Gen. Physiol. 129, 109–120 (2007).
Solaro, C. R. & Lingle, C. J. Trypsin-sensitive, rapid inactivation of a calcium-activated potassium channel. Science 257, 1694–1698 (1992).
Kuo, M. M., Maslennikov, I., Molden, B. & Choe, S. The desensitization gating of the MthK K+ channel is governed by its cytoplasmic amino terminus. PLoS Biol. 6, e223 (2008).
Posson, D. J., Rusinova, R., Andersen, O. S. & Nimigean, C. M. Calcium ions open a selectivity filter gate during activation of the MthK potassium channel. Nat. Commun. 6, 8342 (2015).
Thomson, A. S. et al. Initial steps of inactivation at the K+ channel selectivity filter. Proc. Natl Acad. Sci. USA 111, E1713–E1722 (2014).
Doyle, D. A. et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280, 69–77 (1998).
Zhou, Y., Morais-Cabral, J. H., Kaufman, A. & MacKinnon, R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature 414, 43–48 (2001).
Pau, V. P. et al. Structure and function of multiple Ca2+-binding sites in a K+ channel regulator of K+ conductance (RCK) domain. Proc. Natl Acad. Sci. USA 108, 17684–17689 (2011).
Colquhoun, D. Binding, gating, affinity and efficacy: the interpretation of structure–activity relationships for agonists and of the effects of mutating receptors. Br. J. Pharmacol. 125, 923–947 (1998).
Smith, F. J., Pau, V. P., Cingolani, G. & Rothberg, B. S. Structural basis of allosteric interactions among Ca2+-binding sites in a K+ channel RCK domain. Nat. Commun. 4, 2621 (2013).
Wilkens, C. M. & Aldrich, R. W. State-independent block of BK channels by an intracellular quaternary ammonium. J. Gen. Physiol. 128, 347–364 (2006).
Posson, D. J., McCoy, J. G. & Nimigean, C. M. The voltage-dependent gate in MthK potassium channels is located at the selectivity filter. Nat. Struct. Mol. Biol. 20, 159–166 (2013).
Pau, V. P., Abarca-Heidemann, K. & Rothberg, B. S. Allosteric mechanism of Ca2+ activation and H+-inhibited gating of the MthK K+ channel. J. Gen. Physiol. 135, 509–526 (2010).
Zagotta, W. N., Hoshi, T. & Aldrich, R. W. Restoration of inactivation in mutants of Shaker potassium channels by a peptide derived from ShB. Science 250, 568–571 (1990).
Murrell-Lagnado, R. D. & Aldrich, R. W. Interactions of amino terminal domains of Shaker K channels with a pore blocking site studied with synthetic peptides. J. Gen. Physiol. 102, 949–975 (1993).
Murrell-Lagnado, R. D. & Aldrich, R. W. Energetics of Shaker K channels block by inactivation peptides. J. Gen. Physiol. 102, 977–1003 (1993).
Antz, C. et al. NMR structure of inactivation gates from mammalian voltage-dependent potassium channels. Nature 385, 272–275 (1997).
Bentrop, D., Beyermann, M., Wissmann, R. & Fakler, B. NMR structure of the “ball-and-chain” domain of KCNMB2, the β2-subunit of large conductance Ca2+- and voltage-activated potassium channels. J. Biol. Chem. 276, 42116–42121 (2001).
Schott, M. K., Antz, C., Frank, R., Ruppersberg, J. P. & Kalbitzer, H. R. Structure of the inactivating gate from the Shaker voltage gated K+ channel analyzed by NMR spectroscopy. Eur. Biophys. J. 27, 99–104 (1998).
Wissmann, R. et al. NMR structure and functional characteristics of the hydrophilic N terminus of the potassium channel β-subunit Kvβ1.1. J. Biol. Chem. 274, 35521–35525 (1999).
Mastronarde, D. N. & Serial, E. M. SerialEM: a program for automated tilt series acquisition on Tecnai microscopes using prediction of specimen position. Microsc. Microanal. 9, 1182–1183 (2003).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Zivanov, J., Nakane, T. & Scheres, S. H. W. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Brown, A. et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D 71, 136–153 (2015).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D 74, 531–544 (2018).
Barad, B. A. et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015).
Wang, R. Y. et al. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. eLife 5, e17219 (2016).
The PyMOL Molecular Graphics System (DeLano Scientific, 2002).
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996).
Jones, D. T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 (1999).
Posson, D. J., Rusinova, R., Andersen, O. S. & Nimigean, C. M. Stopped-flow fluorometric ion flux assay for ligand-gated ion channel studies. Methods Mol. Biol. 1684, 223–235 (2018).
Brooks, B. R. et al. CHARMM: the biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 (2009).
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005).
Shaw, D. E. et al. Anton 2: raising the bar for performance and programmability in a special-purpose molecular dynamics supercomputer. In Proceedings of The International Conference for High Performance Computing, Networking, Storage and Analysis 41–53 (IEEE Press, 2014).
Klauda, J. B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010).
MacKerell, A. D., Jr et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 (1998).
Mackerell, A. D., Jr, Feig, M. & Brooks, C. L., III. Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 25, 1400–1415 (2004).
Noskov, S. Y., Bernèche, S. & Roux, B. Control of ion selectivity in potassium channels by electrostatic and dynamic properties of carbonyl ligands. Nature 431, 830–834 (2004).
Andersen, H. C. Molecular dynamics simulations at constant pressure and/or temperature. J. Chem. Phys. 72, 2384–2393 (1980).
Feller, S. E., Zhang, Y., Pastor, R. W. & Brooks, B. R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 103, 4613–4621 (1995).
Martyna, G. J., Tobias, D. J. & Klein, M. L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 101, 4177–4189 (1994).
Nosé, S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 81, 511–519 (1984).
Hoover, W. G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A 31, 1695–1697 (1985).
Andersen, H. C. Rattle: A “velocity” version of the shake algorithm for molecular dynamics calculations. J. Comput. Phys. 52, 24–34 (1983).
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: An N•log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089 (1993).
Acknowledgements
All electron microscopy screening and data collection for the Ca2+-bound MthK wild-type and Δ2–17 structures was performed at the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the Simons Foundation (349247), NYSTAR, and the NIH National Institute of General Medical Sciences (GM103310). The data collection for the apo MthK structure was performed at the CryoEM core facility at UMass. Initial negative stain screening was performed at the Weill Cornell Microscopy and Image Analysis Core Facility, with the help of L. Cohen-Gould. We thank D. Bobe and L. Yen for their technical support during grid screening and L. Yen, M. Kopylov and E. Eng for their support during data collection; C. Xu and K. Song for their support; B. Chanda and Y. Jiang for the gift of the non-inactivating MthK plasmid; S. Scheuring and A. Accardi for critically reading the manuscript; and D. Acehan and C. Boiteux for helpful discussions. This work was supported in part by a National Institutes of Health grant (R01GM088352) to C.M.N., and by the National Institutes of Health (U01-HL126273), National Health and Medical Research Council (APP1104259 and APP1141974), Australian Research Council (DP170101732), D. E. Shaw Anton 2 (PSCA17045P via NIH RC2GM093307), National Computational Initiative, and the Medical Advances Without Animals Trust to T.W.A.
Author information
Authors and Affiliations
Contributions
C.F. and C.M.N. designed the experiments and analysed the results; C.F. acquired cryo-EM data and determined structures of all calcium-bound MthK and Δ2–17 MthK structures; J.R. acquired cryo-EM data and determined the apo MthK channel structure; N.S. performed protein expression and purification, electrophysiology and stopped-flow experiments; E.F. and T.W.A. performed the molecular dynamics simulations; and C.F. and C.M.N. prepared the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Sequence alignments.
a, Sequence alignment of MthK with human Slo1 and Aplysia californica (Ac) Slo1. Secondary structures of closed state MthK are labelled on top. b, Sequence alignment of MthK RCK domain with human Slo1 RCK1 and RCK2 domain. Blue regions indicate similarity and dark blue regions indicate identity. c, Sequence alignment shows inactivation N-terminal from MthK (accession number CEP 36137), Shaker B (accession number CAA 29917), Kv-β1.1 (accession number CAA 50000), BK-β3a (accession number NP_741979) and BK-β2a (accession number NP_001265840). d, Secondary structure prediction (PSIPRED server (see method)) indicates that the N-terminal 17 residues of MthK form a helix. Dashed box indicates the manually built residues in the structure.
Extended Data Fig. 2 Single-particle cryo-EM characterization of closed MthK in the absence of Ca2+.
a, Size-exclusion chromatography (SEC) of MthK reconstituted in nanodiscs composed of 3:1 POPE:POPG lipids with MSP1E3. The main (highest amplitude) peak from SEC was separated using SDS–PAGE (inset), showing the components of MthK nanodiscs (indicated). b, Representative micrograph of the MthK nanodisc sample in 0 Ca2+ and 5 mM EDTA (left). Selected 2D-class averages (right). c, Final cryo-EM map of closed MthK coloured by local resolution. d, FSC curves between the two independently refined half-maps after masking (black curve) and from cross validation between the atomic model, refined against half-map 1, against masked sum of both half-maps (red). e, Angular distribution of particles used in the reconstitution. f, Density of MthK EDTA closed state. Segmented cryo-EM density maps (grey mesh) of closed MthK in the absence of Ca2+. The fitted corresponding atomic model is in cyan.
Extended Data Fig. 3 Fenestrations and C-linker in closed MthK.
a, b, Surface representation of MthK closed (a) and open (PDB 3LDC) (b) state transmembrane domains, coloured by amino acid hydrophobicity. The membrane boundaries are indicated. No fenestration was observed in the open state. c, d, A tunnel, drawn with HOLE, shows how fenestrations (drawn through only two opposing subunits) connect the lipid bilayer with the inside of the cavity (grey). e, The resolved C-linker domain and two extra helical turns of TM2 (blue) shown in one MthK subunit. f, Zoomed-in dashed square in e, showing direct interactions between residues in C-linker (blue) and the RCK N-lobe (green). The neighbouring subunit is in beige. The residues that may contribute to C-linker stabilization via hydrophobic and electrostatic interactions are indicated.
Extended Data Fig. 4 Summary of RCK tilt, TM2 bend, conformational changes and lipid interactions from simulations.
a, Snapshot from one (out of ten simulations) representative simulation showing the maximum RCK tilt. Membrane deformations are indicated by the displacements of lipid phosphates (white spheres). Black dotted line highlights the bend of the front TM2 helix. b, Graphs showing variations in RCK tilt and TM2 bend shown for each of the ten simulations where a positive or negative value represents tilting towards the subunit with TM2 located at the front or back. c, Detail showing the bend of TM2 helices. d, The mean time-lagged cross-correlation of RCK tilt and TM2 bend obtained from analysis of all ten simulations shows how the correlation occurs instantaneously (zero lag) with a value of 0.25 ± 0.05. Error bars represent ± s.e.m.; n = 10 independent simulations. e, Alignment on all four RCK domains. RCK dimers rotated upward (red arrows) within the gating ring. f, Time series of r.m.s.d. of each subunit reveals slow relaxation over 300 ns, increasing owing to the vertical rotations of the subunits as membrane interactions are formed, with asymmetric fluctuations seen during the simulation. g, Time-series r.m.s.d. of individual RCK dimers, revealing some maintained asymmetry. These changes occur owing to changes in loops, although overall the structure is preserved, with lower r.m.s.d. values. All r.m.s.d. errors are between 0.01 and 0.04 Å (not shown). h, Channel structure at the end of a 500-ns simulation showing membrane interactions, indicating acidic and basic residues involved. i, Vertical movements of the C-lobe relative to the membrane (top) are well-correlated with tilting movements, and they lead to increased number of contacts between C-lobes and lipids (bottom), with similar results for N-lobes (not shown). j, There is little change in gate size at L95 (orange) and I99 (purple) during the simulation (top), however, the gate grows rapidly during the first several nanoseconds at Q103 (green). This growth is preceded by the upwards movement of K114 sidechain, positioned on the C-linker (bottom panel). This figure shows representative time series from 1 of the 10 independent simulations performed. See Supplementary Video 4.
Extended Data Fig. 5 Cryo-EM data processing workflow for the MthK Ca2+ dataset.
After refinement, the classes without clear transmembrane density were discarded and marked by X. The classes were classified into four groups according to the tilting degree and coloured according to the class name. Red for highly tilted classes, blue for medium tilted classes, green for mildly tilted classes and yellow for the closed state.
Extended Data Fig. 6 Overview of all the structures obtained from the MthK Ca2+ dataset.
a, The structures with clear transmembrane density were kept; they were classified into four groups according to the tilt angle between the nanodisc (TMD) and RCK ring: highly tilted (red), medium tilted (blue), mildly tilted (green) and closed state (yellow). Finally, there are nine structures in total: three different structures in the highly tilted group (MthK Ca2+ states 1, 1.2 and 1.3), two different structures in the medium tilted group (MthK Ca2+ state 2 and 2.2), three different structures in the mildly tilted group (MthK Ca2+ state 3, 3.2 and 3.3), and one closed state. b, Cryo-EM data processing workflow for only the gating ring structures of the MthK Ca2+ dataset, excluding the closed state. Two different structures were identified by Relion 3D classification (named RCK state 1 and RCK state 2). c, Overlay of RCK states 1 and 2. Slight differences are observed between the two structures.
Extended Data Fig. 7 Single-particle cryo-EM characterization of MthK Ca2+-bound states.
a–d, MthK Ca2+ state 1 (highly tilted). a, Final cryo-EM map coloured by local resolution. b, FSC curve between two independently refined half-maps, after masking. c, Selected 2D-class averages. d, Angular distribution of particles used in the reconstitution. e–h, MthK Ca2+ state 2 (medium-tilted) e, Final cryo-EM map coloured by local resolution. f, FSC curve between two independently refined half-maps, after masking. g, Selected 2D-class averages. h, Angular distribution of particles used in the reconstitution. i–l, MthK Ca2+ state 3 (mildly tilted). i, Final cryo-EM map coloured by local resolution. j, FSC curve between two independently refined half-maps, after masking. k, Selected 2D-class averages. l, Angular distribution of particles used in the reconstitution. m–p, Ca2+-bound closed MthK. m, Final cryo-EM map coloured by local resolution. n, FSC curve between two independently refined half-maps, after masking. o, Selected 2D-class averages from particles. p, Angular distribution of particles used in the reconstitution.
Extended Data Fig. 8 Single-particle cryo-EM characterization of the gating-ring structures in the MthK Ca2+ dataset.
a, Final cryo-EM map of RCK state 1 coloured by local resolution. b, FSC curves between the two independently refined half-maps of MthK Ca2+ RCK state 1 (black), and from cross validation between the atomic model refined against the final cryo-EM map (red). c, Angular distribution of particles used in the reconstitution of RCK state 1. d, Final cryo-EM map of RCK state 2 coloured by local resolution. e, FSC curves between the two independently refined half-maps of MthK Ca2+ RCK state 2 (black), and from cross validation between the atomic model refined against the final cryo-EM map (red). f, Angular distribution of particles used in the reconstitution of RCK state 2. g, Segmented cryo-EM density maps (grey mesh) of RCK state 2. The fitted corresponding atomic model is in cyan.
Extended Data Fig. 9 Functional characteristics of MthK.
a, Single-channel characteristics of MthK(Δ2–17) are similar to the wild type. Top, representative single-channel recording traces from MthK(Δ2–17) in horizontal lipid bilayers made of POPE:POPG (3:1) liposomes in decane at +100 mV without and with 5 mM Ca2+. Traces are filtered at 200 Hz for display. Single-channel current–voltage curves (bottom left) and Po as a function of voltage (bottom right) for MthK(Δ2–17) (red symbols) compared to wild type (dashed black lines). For MthK(WT), data are mean ± s.e.m. of 5 measurements for all membrane potentials except at −100, 75, and 100 mV, which contain 6 measurements. For MthK(Δ2–17), data are mean ± s.e.m. of 5 (−50 mV), 6 (−75, 50 and 125 mV), 7 (−125 mV), 8 (−25 and 75 mV) and 9 (25 and 100 mV) measurements. Each measurement is from a separate bilayer. b, Relative Tl+ flux rates as a function of incubation time of MthK WT (blue) and MthK Δ2–17 (red)-containing POPE:POPG (3:1) liposomes with 5 mM Ca2+. Symbols are the mean ± s.d. from three independent experiments. c, Fluorescence quench curves for MthK(WT)-containing DOPC:POPG (3:1) liposomes after 1 or 10 s (dark and light blue, respectively) incubation with 5 mM Ca2+. Control fluorescence is in the absence of Tl+ (black). A small leak of Tl+ into liposomes was observed in the absence of Ca2+ (light grey) and in the MthK-free liposomes (dark grey). Three experiments were performed with similar results. d, Fluorescence quench curves for MthK(Δ2–17)-containing DOPC:POPG (3:1) liposomes after 1 or 10 s (brown and pink, respectively) incubation with 5 mM Ca2+. The control was performed similarly as that for the experiment with the MthK(WT) liposomes. Experiments were performed three times with similar results. e, f, Simulation snapshots illustrating salt bridges between the basic residues on the N terminus (blue) and the ring of glutamates at the intracellular pore entrance (e), and hydrophobic interactions between the N terminus (blue) and hydrophobic residues lining the pore cavity (f). Only three subunits of the MthK transmembrane domain are shown for clarity. The residues in stick representation are coloured the same as the individual subunits. The calibration bar indicates the position of the COM of the peptide parallel to the channel pore axis (see Methods). g, Convergence of the free energy profile from Umbrella Sampling simulations for the N-terminal peptide plugging the MthK pore (Fig. 4f). Convergence to within 1 kcal mol−1 was achieved in 23 ns.
Extended Data Fig. 10 Single-particle cryo-EM characterization of MthK(Δ2–17) in the presence of Ca2+.
a–c, Cryo-EM map coloured by local resolution, FSC curve between two independently refined half-maps and angular distribution of particles for state 1. d–f, Cryo-EM map coloured by local resolution, FSC curve between two independently refined half-maps and angular distribution of particles for state 2. g–i, Cryo-EM map coloured by local resolution, FSC curve between two independently refined half-maps and angular distribution of particles for state 3. j–l, Cryo-EM map coloured by local resolution, FSC curve between two independently refined half-maps and angular distribution of particles for RCK state 1. m–o, Cryo-EM map coloured by local resolution, FSC curve between two independently refined half-maps and angular distribution of particles for RCK state 2.
Extended Data Fig. 11 Gating-ring assembly in the open MthK structures and pore densities.
a–c, Cryo-EM density map top (a) and side (c) views, and atomic model of the gating ring structure (b). Interfaces and Ca2+ binding sites are indicated. The UP RCK dimers are in red–blue and the DOWN RCK dimers are in green–yellow. d, Cartoon illustrating the RCK dimer packing within the gating ring of the MthK crystal structure (PDB 1LNQ, left, four-fold symmetry) and our MthK open state (right, two-fold symmetry). Illustrations of the assembly interface 1 (e) and assembly interface 2 (f), in which the MthK gating ring from the crystal structure is in beige (PDB 1LNQ), and from the cryo-EM open structure in green, blue, yellow and red. The structures are aligned by blue subunit in (e) and red subunit in (f). g, Transmembrane domains of MthK open state 2 and open state 3. Density maps (grey mesh) from only two subunits are shown with overlaid model in blue and yellow cartoon. The N-terminal plug is in dark blue. h, Transmembrane domains of MthK(Δ2–17) state 2 and state 3. Colours as in g.
Extended Data Fig. 12 The Ca2+-binding sites in the Ca2+-bound closed MthK and RCK state 2 gating ring structures.
a, Overlay of Ca2+-bound closed MthK (red) and Ca2+-free closed MthK (EDTA structure, blue). The two structures are very similar. b, Ca2+ binding sites in the Ca2+-bound closed MthK. Site 1a and 1b are indicated. Density map of Ca2+-binding site 1a (c) and 1b (d). Ca2+ is in orange. The water near the Ca2+ is in red. e, Overview of RCK state 2 structure. Colour scheme is the same as in Fig. 5. The RCK dimer is indicated and shown in detail in f. f, Overlay of the RCK dimer from RCK state 2 detailed in e with the crystal structure of the Ca2+-bound RCK dimer (PDB 4L73, beige). g, The RCK dimer from RCK state 2 with the 6 Ca2+ binding sites indicated. h–m, Density map of Ca2+ binding site 1a (h), 1b (i), 2a (j), 2b (k), 3a (l), 3b (m). Ca2+ is coloured in orange. The water close to Ca2+ is coloured red. Residues forming the binding sites (indicated) are rendered in pink and blue sticks as they originate from adjacent subunits.
Supplementary information
Video 1
: Overall view of the cryo-EM structure of MthK closed state. Each of the subunits is in a different colour. Colours are as in Fig1. The fenestration in the transmembrane region is highlighted.
Video 2
: Conformational change of the MthK intracellular gate upon channel opening. The conformational change was calculated by the morph function in Chimera. Two charged residues E92, E96 were highlighted and shown as ball and stick. These two residues twisted facing the pore upon channel opening.
Video 3
: MD simulation of MthK closed state. A sample all-atom simulation of the closed MthK showing how the gating ring is highly flexible and rocks back and forth during 500 ns. This tilting is accompanied by bending of the TM2 helices (Extended Data Fig.4c). The RCK C-lobe, N-lobe and C-linker interact with the membrane (lipid P atoms represented as white spheres) leading to membrane curvature (Extended Data Fig.4h). As the simulation progresses, RCK domains from all four subunits interact with the membrane, causing upward rotation of each RCK domains (Extended Data Fig.4e-g).
Video 4
: Expansion of the intracellular entryway at Q103 associated with C-linker movement. The MthK C-linker is highly flexible, which causes the gate to expand. During the first few nanoseconds of this sample simulation, the sidechains of K114 and D111 move upwards and interact with lipid PE and PG headgroups, causing the loss of the interaction of D111 with R154 and rapid increase of the gate size, corresponding to an increase in Q103 radial position from 6.1 Å to 8.5 Å (Extended Data Fig.4).
Video 5
: Overall view of the cryo-EM structure of MthK open-inactivated state 1. Each subunit is in a different colour. Colours are as in Fig. 3. The C-linker and the N-terminal inactivation plug (blue) are highlighted.
Video 6
: N-terminus exiting the pore of WT MthK. The pore of MthK is shown in the same colours as in Fig. 4. The initial structure is the cryo-EM structure of the TM domains of MthK Ca2+ state 1 with the N-terminus inside the pore. The side chains of E92, E96 on TM2 and E4, R9, K10, H11 R14, K17 on the N-terminal peptide are shown in order to illustrate the interactions between the pore and the N-terminus.
Video 7
: Conformational change of the RCK gating ring from closed to open state. Each of the subunits is in a different colour. The colour is the same as in Extended Data Fig.11. The conformational change was calculated by the morph function in Chimera.
Video 8
: Morph of subunit A with the ordered C-linker between closed and open-inactivated state. The subunits from two structures were aligned on the transmembrane region. The conformational change was calculated by the morph function in PyMOL.
Rights and permissions
About this article
Cite this article
Fan, C., Sukomon, N., Flood, E. et al. Ball-and-chain inactivation in a calcium-gated potassium channel. Nature 580, 288–293 (2020). https://doi.org/10.1038/s41586-020-2116-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2116-0
This article is cited by
-
Calcium-gated potassium channel blockade via membrane-facing fenestrations
Nature Chemical Biology (2024)
-
Mechanism underlying delayed rectifying in human voltage-mediated activation Eag2 channel
Nature Communications (2023)
-
Central cavity dehydration as a gating mechanism of potassium channels
Nature Communications (2023)
-
Hydrophobic gating in bundle-crossing ion channels: a case study of TRPV4
Communications Biology (2023)
-
N-type fast inactivation of a eukaryotic voltage-gated sodium channel
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.