Abstract
The innate immune regulator STING is a critical sensor of self- and pathogen-derived DNA. DNA sensing by STING leads to the induction of type-I interferons (IFN-I) and other cytokines, which promote immune-cell-mediated eradication of pathogens and neoplastic cells1,2. STING is also a robust driver of antitumour immunity, which has led to the development of STING activators and small-molecule agonists as adjuvants for cancer immunotherapy3. Pain, transmitted by peripheral nociceptive sensory neurons (nociceptors), also aids in host defence by alerting organisms to the presence of potentially damaging stimuli, including pathogens and cancer cells4,5. Here we demonstrate that STING is a critical regulator of nociception through IFN-I signalling in peripheral nociceptors. We show that mice lacking STING or IFN-I signalling exhibit hypersensitivity to nociceptive stimuli and heightened nociceptor excitability. Conversely, intrathecal activation of STING produces robust antinociception in mice and non-human primates. STING-mediated antinociception is governed by IFN-Is, which rapidly suppress excitability of mouse, monkey and human nociceptors. Our findings establish the STING–IFN-I signalling axis as a critical regulator of physiological nociception and a promising new target for treating chronic pain.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Raw data are available from the corresponding author upon request. Source data are provided with this paper.
References
Ishikawa, H. & Barber, G. N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 (2008).
Woo, S. R. et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 41, 830–842 (2014).
Kwon, J. & Bakhoum, S. F. The cytosolic DNA-sensing cGAS–STING pathway in cancer. Cancer Discov. 10, 26–39 (2020).
Julius, D. & Basbaum, A. I. Molecular mechanisms of nociception. Nature 413, 203–210 (2001).
Donnelly, C. R., Chen, O. & Ji, R. R. How do sensory neurons sense danger signals? Trends Neurosci. 43, 822–838 (2020).
Sneddon, L. U. Comparative physiology of nociception and pain. Physiology (Bethesda) 33, 63–73 (2018).
Baral, P., Udit, S. & Chiu, I. M. Pain and immunity: implications for host defence. Nat. Rev. Immunol. 19, 433–447 (2019).
Ji, R. R., Chamessian, A. & Zhang, Y. Q. Pain regulation by non-neuronal cells and inflammation. Science 354, 572–577 (2016).
Zheng, Y. et al. Deep sequencing of somatosensory neurons reveals molecular determinants of intrinsic physiological properties. Neuron 103, 598–616.e7 (2019).
Yaksh, T. L. & Rudy, T. A. Analgesia mediated by a direct spinal action of narcotics. Science 192, 1357–1358 (1976).
Costigan, M., Scholz, J. & Woolf, C. J. Neuropathic pain: a maladaptive response of the nervous system to damage. Annu. Rev. Neurosci. 32, 1–32 (2009).
Wang, K. et al. PD-1 blockade inhibits osteoclast formation and murine bone cancer pain. J. Clin. Invest. 130, 3603–3620 (2020).
Mogil, J. S. Animal models of pain: progress and challenges. Nat. Rev. Neurosci. 10, 283–294 (2009).
Navratilova, E. & Porreca, F. Reward and motivation in pain and pain relief. Nat. Neurosci. 17, 1304–1312 (2014).
Ji, R. R., Donnelly, C. R. & Nedergaard, M. Astrocytes in chronic pain and itch. Nat. Rev. Neurosci. 20, 667–685 (2019).
Hummel, M., Lu, P., Cummons, T. A. & Whiteside, G. T. The persistence of a long-term negative affective state following the induction of either acute or chronic pain. Pain 140, 436–445 (2008).
Agarwal, N., Offermanns, S. & Kuner, R. Conditional gene deletion in primary nociceptive neurons of trigeminal ganglia and dorsal root ganglia. Genesis 38, 122–129 (2004).
Haag, S. M. et al. Targeting STING with covalent small-molecule inhibitors. Nature 559, 269–273 (2018).
Usoskin, D. et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci 18, 145–153 (2015).
Fensome, A. et al. Dual inhibition of TYK2 and JAK1 for the treatment of autoimmune diseases: discovery of ((S)-2,2-difluorocyclopropyl)((1R,5S)-3-(2-((1-methyl-1H-pyrazol-4-yl)amino)pyrimidin-4-yl)-3,8-diazabicyclo[3.2.1]octan-8-yl)methanone (PF-06700841). J. Med. Chem. 61, 8597–8612 (2018).
Harari, D. et al. Enhanced in vivo efficacy of a type I interferon superagonist with extended plasma half-life in a mouse model of multiple sclerosis. J. Biol. Chem. 289, 29014–29029 (2014).
Barragán-Iglesias, P. et al. Type I interferons act directly on nociceptors to produce pain sensitization: implications for viral infection-induced pain. J. Neurosci. 40, 3517–3532 (2020).
Todd, A. J. Neuronal circuitry for pain processing in the dorsal horn. Nat. Rev. Neurosci. 11, 823–836 (2010).
Sun, L., Wu, J., Du, F., Chen, X. & Chen, Z. J. Cyclic GMP–AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013).
Ivashkiv, L. B. & Donlin, L. T. Regulation of type I interferon responses. Nat. Rev. Immunol. 14, 36–49 (2014).
Binshtok, A. M., Bean, B. P. & Woolf, C. J. Inhibition of nociceptors by TRPV1-mediated entry of impermeant sodium channel blockers. Nature 449, 607–610 (2007).
Hu, E., Calò, G., Guerrini, R. & Ko, M. C. Long-lasting antinociceptive spinal effects in primates of the novel nociceptin/orphanin FQ receptor agonist UFP-112. Pain 148, 107–113 (2010).
Sjöström, S., Tamsen, A., Persson, M. P. & Hartvig, P. Pharmacokinetics of intrathecal morphine and meperidine in humans. Anesthesiology 67, 889–895 (1987).
Crow, M. K., Olferiev, M. & Kirou, K. A. Type I interferons in autoimmune disease. Annu. Rev. Pathol. 14, 369–393 (2019).
Neufeld, N. J., Elnahal, S. M. & Alvarez, R. H. Cancer pain: a review of epidemiology, clinical quality and value impact. Future Oncol. 13, 833–841 (2017).
Sivick, K. E. et al. Magnitude of therapeutic STING activation determines CD8+ T cell-mediated anti-tumor immunity. Cell Rep. 25, 3074–3085.e5 (2018).
Tan, Y. S. et al. Mitigating SOX2-potentiated immune escape of head and neck squamous cell carcinoma with a STING-inducing nanosatellite vaccine. Clin. Cancer Res. 24, 4242–4255 (2018).
Scheu, S., Dresing, P. & Locksley, R. M. Visualization of IFNβ production by plasmacytoid versus conventional dendritic cells under specific stimulation conditions in vivo. Proc. Natl Acad. Sci. USA 105, 20416–20421 (2008).
Sauer, J. D. et al. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect. Immun. 79, 688–694 (2011).
Jin, L. et al. MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic-di-AMP and cyclic-di-GMP. J. Immunol. 187, 2595–2601 (2011).
Prigge, J. R. et al. Type I IFNs act upon hematopoietic progenitors to protect and maintain hematopoiesis during pneumocystis lung infection in mice. J. Immunol. 195, 5347–5357 (2015).
Schoggins, J. W. et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505, 691–695 (2014).
Sisignano, M. et al. Targeting CYP2J to reduce paclitaxel-induced peripheral neuropathic pain. Proc. Natl Acad. Sci. USA 113, 12544–12549 (2016).
Chen, G. et al. PD-L1 inhibits acute and chronic pain by suppressing nociceptive neuron activity via PD-1. Nat. Neurosci. 20, 917–926 (2017).
Decosterd, I. & Woolf, C. J. Spared nerve injury: an animal model of persistent peripheral neuropathic pain. Pain 87, 149–158 (2000).
Chaplan, S. R., Bach, F. W., Pogrel, J. W., Chung, J. M. & Yaksh, T. L. Quantitative assessment of tactile allodynia in the rat paw. J. Neurosci. Methods 53, 55–63 (1994).
Chen, G., Park, C. K., Xie, R. G. & Ji, R. R. Intrathecal bone marrow stromal cells inhibit neuropathic pain via TGF-β secretion. J. Clin. Invest. 125, 3226–3240 (2015).
Bouet, V. et al. The adhesive removal test: a sensitive method to assess sensorimotor deficits in mice. Nat. Protoc. 4, 1560–1564 (2009).
King, T. et al. Unmasking the tonic-aversive state in neuropathic pain. Nat. Neurosci. 12, 1364–1366 (2009).
Zhang, Z. et al. Persistent pain facilitates response to morphine reward by downregulation of central amygdala GABAergic function. Nat. Neurosci. 39, 2263–2271 (2014).
Ren, B. X. et al. Intrathecal injection of metabotropic glutamate receptor subtype 3 and 5 agonist/antagonist attenuates bone cancer pain by inhibition of spinal astrocyte activation in a mouse model. Anesthesiology 116, 122–132 (2012).
Demaria, O. et al. STING activation of tumor endothelial cells initiates spontaneous and therapeutic antitumor immunity. Proc. Natl Acad. Sci. USA 112, 15408–15413 (2015).
Xu, Z. Z. et al. Inhibition of mechanical allodynia in neuropathic pain by TLR5-mediated A-fiber blockade. Nat. Med. 21, 1326–1331 (2015).
Wang, Z. et al. Anti-PD-1 treatment impairs opioid antinociception in rodents and nonhuman primates. Sci. Transl. Med. 12, eaaw6471 (2020).
Jiang, C. et al. PD-1 Regulates GABAergic neurotransmission and GABA-mediated analgesia and anesthesia. iScience 23, 101570 (2020).
Acknowledgements
We thank J. P.-Y. Ting and B. D. X. Lascelles for helpful discussions and R. Kuner for providing Nav1.8-cre mice. This study was supported by Duke University Anesthesiology research funds and Duke Microbiome Center fund granted to R.-R.J. C.R.D. was also supported by the International Association for the Study of Pain John J. Bonica Trainee Fellowship, NIH T32 training grant GM008600, and the Department of Anesthesiology, Duke University. M.-C.K. and the monkey study was supported by NIH R21 grant DA044450.
Author information
Authors and Affiliations
Contributions
C.R.D. and R.-R.J. conceived and shaped the overall direction of the project. C.R.D., C.J., K.W., A.S.A., Z.W., J.Z., X.L. and M.S.L. performed experiments. H.D. performed behavioural testing in monkeys under the guidance of M.-C.K. Y.L.L. and W.M. contributed to the experimental design. Illustrations were created by C.R.D. using BioRender. C.R.D. and R.-R.J. wrote the manuscript with feedback and discussions from all co-authors.
Corresponding authors
Ethics declarations
Competing interests
R.-R.J. is a consultant of Boston Scientific and received research support from the company unrelated to the present study. C.R.D. and R.-R.J. have also filed a patent, ‘Treatment of Neuropathic and Cancer Pain Using Agonists of Stimulator of Interferon Response cGAMP Interactor-1 (STING1)’ in association with Duke University. The other authors declare no competing interests.
Additional information
Peer review information Nature thanks Bruce Bean, Marzia Malcangio and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 STING is expressed in nociceptive sensory neurons in the DRG.
a, Sting1 (STING) mRNA expression in sensory neuron populations recently profiled and previously described9. Peptidergic nociceptive sensory neurons exhibit the highest expression of STING. b, c, In situ hybridization of Sting1 in adult DRG sensory neurons using RNAscope, in conjunction with Nissl staining to label all neurons (b). STING expression is primarily observed in small-diameter sensory neurons. Quantification of DRG neurons expressing STING (≥5 puncta) or lacking STING found that approximately 60% of DRG neurons express STING. Scale bar is 100 μm. c, Quantification of somal diameter in STING+ and STING- neurons indicated that STING-expressing neurons are primarily small-diameter sensory neurons. d, Schematic indicating the various stimuli which activate STING and its downstream pathway. See Supplementary Information for complete sample sizes, sex, and statistical information.
Extended Data Fig. 2 Antinociceptive effects of STING agonists in naive mice and in mouse models of neuropathic and cancer pain.
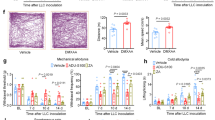
a, Naive mice were administered vehicle or the STING agonist DMXAA via i.t. injection (red arrows), followed by Von Frey testing to determine mechanical thresholds at 4 h following the first (day 1, D1) or second (D2) injection. STING agonists induced a dose-dependent increase in paw withdrawal thresholds, which was further amplified by multiple injections. 10 μg (35 nmol) exhibited the largest effect, and therefore, was used throughout the rest of the study. BL, baseline. b, Naive mice were administered vehicle or ADU-S100, a STING agonist with cross-species activity, via i.t. injection (red arrows) and tested as in a. 25 μg (35 nmol) exhibited the largest increase in paw withdrawal thresholds and this dose was used throughout the rest of the study. c, Systemic administration of DMXAA and ADU-S100 increased paw withdrawal threshold in naive mice for up to 24 h. d, In the CIPN model, i.p. DMXAA and ADU-S100 suppressed mechanical allodynia for up to 48 h. Some toxicity was observed with systemic administration in the CIPN model, as 3 mice in the DMXAA group died 24 h after the second injection. No mice died in the vehicle or ADU-S100 groups. e, A chemotherapy-induced peripheral neuropathy (CIPN) model of neuropathic pain was established with paclitaxel. f, g, In the CIPN model, i.t. DMXAA and ADU-S100 suppressed mechanical allodynia (f) and cold allodynia (g), as determined by response duration (in seconds) to acetone. h, Intrathecal administration with the natural STING ligand 3′,3′-cGAMP also reduced CIPN-induced mechanical allodynia. i, STING agonists were also tested in the sciatic nerve chronic constriction injury (CCI) model of neuropathic pain. j, Intrathecal treatment with DMXAA and ADU-S100 led to prolonged inhibition of mechanical allodynia (as determined by withdrawal threshold). k, l, Administration of DMXAA and ADU-S100 also suppressed cold allodynia (k) in the BCP model. These effects were not secondary to direct antitumour effects, as tumour burden was unaffected by STING agonist treatment and treatment groups still show significant tumour growth (l). m–o, Effects of naloxone on morphine-, DMXAA-, and ADU-S100-induced antinociception. Naloxone (10 mg/kg, i.p.) reversed morphine (2 nmol i.t.)-induced antinociception (m) but had no effect on the antinociceptive effects of DMXAA (35 nmol, i.t.) (n) or ADU-S100 (35 nmol, i.t.) (o). p, Repeated administration of DMXAA (35 nmol, i.t.) did not induce tolerance in naive mice. q–t, Pain and glial reaction after spared nerve injury (SNI). q, Paradigm of experimental design. Pain behaviours and spinal dorsal horn glial cell reaction were assessed following repeated administration with vehicle or DMXAA (red arrows) in mice receiving SNI. r, s, Mechanical allodynia (r) and cold allodynia (s) were significantly reduced in DMXAA-treated mice at later time points (starting at D12). t, Quantification of GFAP+ (astrocytes), Iba1+ (microglia), or DAPI (all cells) in the spinal dorsal horn (SDH) of mice at D21 after sustained vehicle or DMXAA treatment as indicated in q. SNI increased astrocyte activation (GFAP) in vehicle- but not DMXAA-treated mice. Values represent the ipsilateral (injured) SDH normalized to the contralateral (uninjured) SDH. All data are expressed as the mean ± s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001. Statistical comparisons were conducted with two-way ANOVA with Dunnett’s (a–d, f, g, k), Sidak’s (h, j, l, s), Bonferroni’s (m–p, r, t), or Tukey’s post hoc test (m–o). See Supplementary Information for complete sample sizes, sex, and statistical information.
Extended Data Fig. 3 STING agonists induce conditioned place preference in mice with neuropathic pain but not in naive mice.
a, Schematic indicating the experimental setup for the conditioned place preference (CPP) assay used to measure ongoing pain in mice. b, Schematic and timeline indicating CPP protocol for naive mice. CPP in naive mice was performed using repeated pairings (3 trials). c, Repeated pairing with i.t. morphine (2 nmol), but not DMXAA or ADU-S100 (35 nmol), induces CPP in naive mice. d, Schematic and timeline indicating CPP protocol for mice in the chemotherapy-induced peripheral neuropathy (CIPN) model of neuropathic pain. e, A single pairing with i.t. DMXAA and ADU-S100 (35 nmol) induced comparable CPP as clonidine (35 nmol), a strong analgesic when administered via i.t. injection. f, Schematic depicting patch clamp recordings in small diameter dissociated DRG neurons from STING+/+ (gWT) or STINGgt/gt (gKO) mice. g, h, STINGgt/gt mice exhibit increased excitability, as determined by number of action potentials evoked per current step (in 10 pA increments), as indicated by traces (g) and quantification (h). i, j, Nociceptors from STINGgt/gt mice also exhibit decreased rheobase at baseline, as indicated by traces (i) and quantification (j). All data are expressed as the mean ± s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001. Statistical comparisons were conducted with one-way ANOVA with Fisher’s LSD test (c, e), two-way ANOVA with Bonferroni’s post hoc test (h), or two-tailed t-test (j). See Supplementary Information for complete sample sizes, sex, and statistical information.
Extended Data Fig. 4 Mice lacking STING exhibit normal innervation patterns, sensory neuron numbers, and sensorimotor behaviours.
a–f, Hindpaw skin immunostaining for nerve fibres (a, c, e) and their quantification (b, d, f) from STING+/+ (gWT) or STINGgt/gt (gKO) mice. TuJ1 to label all nerve fibres (a), CGRP to label fibres from peptidergic nociceptors (c), or IB4 to label fibres from nonpeptidergic nociceptors (e). In each case, sections were counterstained with DAPI to identify the dermis-epidermis junction. Quantification of TuJ1+ (b), CGRP+ (d) and IB4+ nerve fibres (f) indicate that STING-WT and -gKO mice have similar peripheral innervation densities. Scale bar for (a-f) is 50 μm. g–i, L3-L5 spinal cord segments were collected from STING+/+ and STINGgt/gt mice and immunostained for CGRP (red) and IB4 (green) to label central nociceptive terminals. g, Representative images, scale bar is 100 μm. Image J quantification of CGRP (h) and IB4 (i) pixel density in the spinal dorsal horn (displayed in arbitrary units, A.U.) indicates that there are no changes in central innervation density in STINGgt/gt mice. j, k, Analysis of total DRG neuron numbers in STING+/+ and STINGgt/gt mice. j, L5 DRGs were collected from STING+/+ and STINGgt/gt mice, serially sectioned at 14 μm, and every 3rd section was immunostained for the pan-neuronal marker TuJ1 (purple), counterstained with DAPI (blue). Scale bar is 200 μm. k, Quantification of total DRG neuron numbers indicated that STING+/+ and STINGgt/gt have similar sensory neuron numbers. l–n, No differences were observed between STING+/+ and STINGgt/gt mice in the tape test (l), a measure of sensorimotor function, in an accelerating rotarod test to measure motor function (m), or in locomotor activity in the open field test (30 min test duration) (n). All data are expressed as the mean ± s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001. Statistical comparisons were conducted with two-tailed t-test. See Supplementary Information for complete sample sizes, sex, and statistical information.
Extended Data Fig. 5 STING signalling in nociceptors is required for the antinociceptive effects of STING agonists.
a, Schematic showing mechanism of action of the small molecule STING inhibitors H-151 and C-176. b–d, i.t. administration of H-151 and C-176 induced transient mechanical (b, c) and cold hypersensitivity (d) in naive mice. e, DMXAA and ADU-S100 (35 nmol each, i.t.) increased paw withdrawal thresholds in STING+/+ (gWT) but not in STINGgt/gt (gKO) mice. f, Administration of ADU-S100 (35 nmol, i.t.) did not alter paw withdrawal frequency to a low-threshold 0.16g Von Frey filament in STINGgt/gt mice. g, DMXAA and ADU-S100 increased paw withdrawal threshold in Stingfl/fl (cWT) mice, but no significant effects were observed in Stingfl/fl;Nav1.8-Cre (cKO) mice. A non-statistically significant increase in withdrawal thresholds was observed at later time points (24 h, 48 h) in Stingfl/fl;Nav1.8-Cre mice. h, DMXAA and ADU-S100 (35 nmol each, i.t.) reduced mechanical hypersensitivity (determined by withdrawal frequency to a 0.16 g Von Frey filament) in Stingfl/fl;Nav1.8-Cre mice at 24 h and 48 h. i, Schematic indicating method of resiniferatoxin (RTX) treatment and the effects of STING agonists in RTX-treated mice. j, RTX increased withdrawal latency in the hotplate test (cutoff time = 40 s). k, The early (1 h, 4 h, 24 h) antinociceptive effects of DMXAA and ADU-S100 (35 nmol, i.t.) were abolished by RTX. l, The antinociceptive effects of DMXAA and ADU-S100 (35 nmol, i.t.) showed a normal time course of effects (relative to Fig. 1b) in Prkdcscid mice, which lack mature B and T cells. m, Schematic indicating possible upstream activators of STING in sensory neurons, and proposed mechanism by which STING+ neurons regulate nociception through induction of cytokines and chemokines. While our data support that STING signalling in nociceptors contributes to the early antinociceptive effects of STING agonists, additional cell types including other sensory neuron populations and peripheral immune cells may also contribute to the prolonged antinociceptive effects at later time points. All data are expressed as the mean ± s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001. Statistical comparisons were conducted with two-way ANOVA with Dunnett’s (vs. BL; b–h) or Tukey’s (k, l) post hoc tests. Comparisons between two groups (j) were conducted with two-tailed t-test. See Supplementary Information for complete sample sizes, sex, and statistical information.
Extended Data Fig. 6 STING agonists induce IFN-I production in sensory neurons in vitro and in vivo.
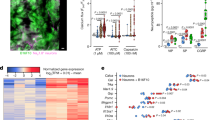
a, Intrathecal administration of STING agonists increases IFN-α in serum 24 h following injection in WT, but not STINGgt/gt mice. b, While basal IFN-β could be detected in DRG tissue from all genotypes, STINGgt/gt mice exhibited significantly lower IFN-β levels. c, d, ADU-S100 treatment of high density DRG neuron cultures from STING+/+ (gWT) or STINGgt/gt (gKO) mice. In WT DRG neurons, ADU-S100 induced release of IFN-α (c) and IFN-β (d) into the culture medium, determined by ELISA. e -h, IFN-β expression in mouse DRG neurons. e, Schematic showing experimental design to determine which cell types within the DRG produce IFN-β in response to i.t. STING activation with ADU-S100 using an Ifnb1YFP reporter mouse. f–h, Administration of ADU-S100 induced an increase in YFP expression (indicating IFN-β expressing cells) within the DRG. f, Representative images showing YFP (purple), TuJ1 (green), and DAPI (blue). Scale bar is 50 μm. g, ADU-S100 increased the proportion of total neurons expressing YFP+ in Ifnb1YFP/YFP mice; virtually no YFP+ cells detected in WT Ifnb1+/+ mice. h, YFP expression was primarily detected in TuJ1+ neurons rather than TuJ1− non-neuronal cell types, although this neuronal expression bias was slightly reduced by ADU-S100 treatment. i–k, Mice lacking Ifnar1 globally (Ifnar1−/−) or selectively in sensory neurons (Ifnar1fl/fl;Nav1.8-Cre) exhibit significantly increased sensitivity to mechanical stimuli determined by paw withdrawal threshold (i) or paw withdrawal frequency (j) to a low stimulus 0.16 g Von Frey filament, compared to their littermate controls. k, These mice also exhibit increased sensitivity to cold stimulation. l, m, CPA testing was performed as in Fig. 1l. Pairing in the preferred chamber with 0.04 g filament induced CPA in Ifnar1−/− mice, but not Ifnar1+/+ littermate control mice. l, Representative trackplots of mouse movement pre- and post-pairing. m, Quantification of CPA score (Pre – post, in seconds). n–q, Action potentials (n, o) and rheobases (p, q) in DRG nociceptors as in Fig. 2h–k. Ifnar1−/− mice exhibit increased excitability, as determined by (n, o) number of action potentials evoked per current step (in 10 pA increments) or (p, q) basal rheobase. r, s, Input resistance was calculated from patch clamp recordings from DRG nociceptors from mice of the indicated genotypes. Increased input resistance was observed in neurons from both Ifnar1−/− (r) and STINGgt/gt (s) mice compared to their WT littermates. All data are expressed as the mean ± s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001. Statistical comparisons were conducted with one-way ANOVA with Tukey’s post hoc test (b, g, i–k), two-way ANOVA with Dunnett’s post hoc test (a, c, d, o), or two-tailed t-test (h, m, q–s). See Supplementary Information for complete sample sizes, sex, and statistical information.
Extended Data Fig. 7 Mice lacking Ifnar1 exhibit normal innervation patterns, sensory neuron numbers, and sensorimotor behaviours.
a–f, Hindpaw skin was collected from Ifnar1+/+ or Ifnar1−/− mice and immunostained for TuJ1 (a), CGRP (c) or IB4 (e) with DAPI as in Extended Data Fig. 4. Quantification of epidermal TuJ1+ (b), CGRP+ (d) and IB4+ (f) nerve fibres indicate that similar skin innervation densities for each marker in WT and KO mice. Scale bar for (a–f) is 50 μm. g–i, L3–L5 spinal cord segments were collected from Ifnar1+/+ or Ifnar1−/− mice and immunostained for CGRP (red) and IB4 (green) to label central nociceptive terminals in the dorsal horn. g, Representative images, scale bar is 100 μm. Image J quantification of CGRP (h) and IB4 (i) pixel density (displayed in arbitrary units, A.U.) indicates similar densities of central innervation. j, k, L5 DRGs were collected from Ifnar1+/+ and Ifnar1−/− mice and total cell counts were performed as in Extended Data Fig. 4 using TuJ1 (purple) and DAPI (blue) (j). Scale bar is 200 μm. k, Quantification of total DRG neuron numbers indicated that Ifnar1+/+ or Ifnar1−/− mice have similar numbers of L5 DRG neurons. l–n, No differences were observed between Ifnar1+/+ and Ifnar1−/− mice in the tape test (l), in the accelerating rotarod test (m), or in locomotor activity in the open field test (n) (30 min duration). All data are expressed as the mean ± s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001. Statistical comparisons were conducted with two-tailed t-test. See Supplementary Information for complete sample sizes, sex, and statistical information.
Extended Data Fig. 8 Type-I interferons regulate nociception in mice via Tyk2.
a, b, Inhibition of endogenous IFN-I signalling via i.t. administration of an anti-IFN-α neutralizing antibody (a) or an anti-IFN-β neutralizing antibody (b) (vs. IgG control, 300 ng) induced transient mechanical allodynia in naive mice. c, Inhibition of the IFN-I signalling adaptor Tyk2 with PF-06700841 (i.t., 1 μg) induced transient, dose-dependent mechanical allodynia in naive mice. d–f, i.t. injection of recombinant murine IFN-α (d), murine IFN-β (produced in mammalian cells) (e), or universal IFN-I (f) increased paw withdrawal thresholds in naive mice. Notably, 100 U exhibited the greatest effects for each recombinant ligand. At higher concentrations some mice exhibited mechanical hypersensitivity. g, Schematic showing downstream IFN-I signalling pathways and the pharmacologic inhibitors used to target Tyk2, PI3K, or MAPKK. h, Pretreatment of naive mice with the Tyk2 inhibitor PF-06700841 (i.t., 1 μg) abolished IFN-β-induced antinociception. i, j, Pre-treatment with 1 μg i.t. U0126, a MAPKK (MEK) inhibitor (i) or 1 μg LY294002, a PI3K inhibitor (j), failed to affect IFN-β-induced increased paw withdrawal threshold. k, Intraplantar (i.pl., for example, hindpaw) administration of IFN-α at high concentrations induced prolonged mechanical hypersensitivity. l, i.pl. injection of 300U IFN-α evokes mechanical hypersensitivity in both WT and Ifnar1−/− mice, as determined by paw withdrawal frequency to repeated stimulation with a very low threshold Von Frey filament (0.04 g, selected due to the baseline hypersensitivity of Ifnar1−/− mice). m, Mechanical hypersensitivity induced by i.pl. injection of 300U IFN-α is attenuated by i.t. IFN-α administration, as determined by paw withdrawal threshold (left panel) or withdrawal frequency (right panel) to repeated stimulation with 0.16g Von Frey filament. All data are expressed as the mean ± s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001. Statistical comparisons were conducted with two-way ANOVA with Bonferroni’s (a, b, m) or Dunnett’s post hoc test (c–f, h–l). See Supplementary Information for complete sample sizes, sex, and statistical information.
Extended Data Fig. 9 Peripheral and central actions of STING-mediated IFN-I signalling in DRG and spinal cord.
a, L1-L5 DRGs were isolated from STINGfl/fl (cWT), STINGfl/fl;Nav1.8-Cre (cKO), and STINGgt/gt (gKO) mice and incubated ex vivo with vehicle (left DRGs) or 30 μM ADU-S100 (right DRGs) for 2h, followed by lysis and IFN-α and IFN-β ELISA. b, c, IFN-α (b) and IFN-β (c) levels in DRG lysate were increased by ex vivo incubation with ADU-S100 in WT mice. ADU-S100 induced a significant elevation of IFN-β in STINGfl/fl;Nav1.8-Cre DRG lysate, but this increase was significantly lower than that seen in WT mice. d, e, DRGs from naive mice were incubated ex vivo in control IgG or anti-IFNβ for 2h followed by whole-mount patch clamp recordings from DRG nociceptors. Representative traces (d) and quantification (e) of current-evoked action potentials. Anti-IFNβ treatment significantly increased action potential firing. f–h, Recordings of miniature EPSCs (mEPSCs) from outer lamina II spinal dorsal horn neurons in spinal cord slices from mice of the indicated genotypes. Representative traces (f) and quantification of mEPSC frequency (g) and amplitude (h). Ifnar1−/− mice exhibit increased frequency and amplitude compared to WT littermates. No significant increase was observed in Ifnar1fl/fl;Nav1.8-Cre mice relative to Ifnar1fl/fl controls. i, Representative images of RNAscope showing Sting1 (STING) mRNA (red) in conjunction with Iba1 immunostaining to label microglia (green) in the spinal cord dorsal horn (SDH). Yellow arrows indicate STING+/Iba1+ cells. Scale bar represents 20 μm. j, Quantification indicates that STING is predominantly expressed by microglia in the SDH. k, l, IFN-α (k) and IFN-β (l) levels in SDH lysate in mice of the indicated genotypes 24 h after STING agonist administration showing induction of IFN-α but not IFN-β. Basal IFN-β production was significantly lower in STINGgt/gt mice. m–o. Ex vivo incubation of spinal cord slices from naive mice with ADU-S100. Representative traces (m) and quantification of frequency (n) and amplitude (o) of spontaneous EPSCs (sEPSCs) from outer lamina II spinal dorsal horn neurons. ADU-S100 significantly reduced the frequency and amplitude of sEPSCs. p–r, Acute perfusion of spinal cord slices with IFN-I and recording of TTX-resistant mEPSCs from outer lamina II spinal dorsal horn neurons. p, Representative traces from Ifnar1+/+ and Ifnar1−/− mice, with IFN-I perfused as indicated and the areas indicated by the black bars magnified for enhanced resolution. Quantification of frequency (q) and amplitude (r) of mEPSC in neurons from WT and KO mice pre- and 1 min post- perfusion with IFN-I. Acute IFN-I treatment significantly reduced the frequency and amplitude of mEPSCs in WT mice. All data are expressed as the mean ± s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001. Statistical comparisons were conducted with one-way or two-way ANOVA with Sidak’s (b, c, g, h) or Bonferroni post hoc test (e, k, l, q, r) or two-tailed t-test (n, o). See Supplementary Information for complete sample sizes, sex, and statistical information.
Extended Data Fig. 10 dsDNA induces antinociception via the cGAS/STING/IFN-I pathway and proposed mechanism of STING/IFN-I regulation of nociception.
a, i.t. IFN-α and IFN-β elevated mechanical thresholds in WT mice and could transiently rescue the mechanical hypersensitivity phenotype of STINGgt/gt mice back to levels comparable to WT baseline levels. b, Schematic illustrating intracellular poly(dA:dT) induces cGAS/STING-dependent IFN-I upregulation whereas intracellular poly(I:C) induces IFN-I response through a STING-independent mechanism. c, d, Mice of the indicated genotypes were administered 1 μg poly(I:C) or 1 μg poly(dA:dT) via i.t. injection followed by Von Frey testing to determine mechanical thresholds at the indicated time points. c, Intrathecal poly(I:C) elevated mechanical thresholds in Ifnar1fl/fl (WT), STINGgt/gt, and cGAS−/− mice, but these effects were abolished in Ifnar1fl/fl;Nav1.8-Cre and RIG-I−/− mice. d, Intrathecal poly(dA:dT) elevated mechanical thresholds in WT and RIG-I−/− mice for up to 24 h, but these effects were abolished in Ifnar1fl/fl;Nav1.8-Cre, STINGgt/gt, and cGAS−/− mice. e–g, Sodium currents were recorded from DRG nociceptors cultured from Ifnar1+/+ or Ifnar1−/− mice, perfused with vehicle or IFN-I for 2 min as indicated. Representative traces (e) and quantification (f) of Na+ currents demonstrating that rIFN-I treatment led to a reduction in sodium currents. g, Nociceptors from Ifnar1-gKO mice exhibited increased Na+ current amplitude at baseline. h, i, HEK293 cells stably expressing the voltage-gated sodium channel Nav1.7 were perfused as in panel f and Na+ currents were recorded. Representative traces (h) and Timecourse (i) of Nav1.7-mediated currents. IFN-I perfusion reduced Nav1.7-mediated currents. All data are expressed as the mean ± s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001. Statistical comparisons were conducted with two-way ANOVA with Dunnett’s post hoc test (a, c, d), two-way ANOVA with Bonferroni’s post hoc test (f, i), or two-tailed t-test (g). See Supplementary Information for complete sample sizes, sex, and statistical information.
Supplementary information
Supplementary Table
This file contains Supplementary Table 1, which describes the complete list of genotypes, treatment groups, animal sex, and total animal numbers for each figure.
Source data
Rights and permissions
About this article
Cite this article
Donnelly, C.R., Jiang, C., Andriessen, A.S. et al. STING controls nociception via type I interferon signalling in sensory neurons. Nature 591, 275–280 (2021). https://doi.org/10.1038/s41586-020-03151-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-03151-1
This article is cited by
-
Endophilin A2 controls touch and mechanical allodynia via kinesin-mediated Piezo2 trafficking
Military Medical Research (2024)
-
Direct paraventricular thalamus-basolateral amygdala circuit modulates neuropathic pain and emotional anxiety
Neuropsychopharmacology (2024)
-
IL-33/ST2 antagonizes STING signal transduction via autophagy in response to acetaminophen-mediated toxicological immunity
Cell Communication and Signaling (2023)
-
Mechanism and therapeutic potential of targeting cGAS-STING signaling in neurological disorders
Molecular Neurodegeneration (2023)
-
STING mediates experimental osteoarthritis and mechanical allodynia in mouse
Arthritis Research & Therapy (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.