Abstract
Epithelial-to-mesenchymal transitions (EMTs) are phenotypic plasticity processes that confer migratory and invasive properties to epithelial cells during development, wound-healing, fibrosis and cancer1,2,3,4. EMTs are driven by SNAIL, ZEB and TWIST transcription factors5,6 together with microRNAs that balance this regulatory network7,8. Transforming growth factor β (TGF-β) is a potent inducer of developmental and fibrogenic EMTs4,9,10. Aberrant TGF-β signalling and EMT are implicated in the pathogenesis of renal fibrosis, alcoholic liver disease, non-alcoholic steatohepatitis, pulmonary fibrosis and cancer4,11. TGF-β depends on RAS and mitogen-activated protein kinase (MAPK) pathway inputs for the induction of EMTs12,13,14,15,16,17,18,19. Here we show how these signals coordinately trigger EMTs and integrate them with broader pathophysiological processes. We identify RAS-responsive element binding protein 1 (RREB1), a RAS transcriptional effector20,21, as a key partner of TGF-β-activated SMAD transcription factors in EMT. MAPK-activated RREB1 recruits TGF-β-activated SMAD factors to SNAIL. Context-dependent chromatin accessibility dictates the ability of RREB1 and SMAD to activate additional genes that determine the nature of the resulting EMT. In carcinoma cells, TGF-β–SMAD and RREB1 directly drive expression of SNAIL and fibrogenic factors stimulating myofibroblasts, promoting intratumoral fibrosis and supporting tumour growth. In mouse epiblast progenitors, Nodal–SMAD and RREB1 combine to induce expression of SNAIL and mesendoderm-differentiation genes that drive gastrulation. Thus, RREB1 provides a molecular link between RAS and TGF-β pathways for coordinated induction of developmental and fibrogenic EMTs. These insights increase our understanding of the regulation of epithelial plasticity and its pathophysiological consequences in development, fibrosis and cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Source Data for Fig. 2d, i, j and Extended Data Figs. 2a, d, 3b, c, e, 4c, d, 5c, i, 6d, k, 7c are provided with the online version of the paper. The ChIP–seq and RNA-seq data have been deposited in the Gene Expression Omnibus under accession numbers GSE118765 and GSE128958. All other data are available from the authors on reasonable request.
Change history
15 January 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41586-020-1956-y
References
Arnold, S. J. & Robertson, E. J. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 10, 91–103 (2009).
Dongre, A. & Weinberg, R. A. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 20, 69–84 (2019).
Ferretti, E. & Hadjantonakis, A. K. Mesoderm specification and diversification: from single cells to emergent tissues. Curr. Opin. Cell Biol. 61, 110–116 (2019).
Nieto, M. A., Huang, R. Y., Jackson, R. A. & Thiery, J. P. EMT: 2016. Cell 166, 21–45 (2016).
Batlle, E. et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2, 84–89 (2000).
Cano, A. et al. The transcription factor snail controls epithelial–mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2, 76–83 (2000).
Brabletz, S. & Brabletz, T. The ZEB/miR-200 feedback loop—a motor of cellular plasticity in development and cancer? EMBO Rep. 11, 670–677 (2010).
De Craene, B. & Berx, G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer 13, 97–110 (2013).
David, C. J. & Massagué, J. Contextual determinants of TGFβ action in development, immunity and cancer. Nat. Rev. Mol. Cell Biol. 19, 419–435 (2018).
Heldin, C. H., Vanlandewijck, M. & Moustakas, A. Regulation of EMT by TGFβ in cancer. FEBS Lett. 586, 1959–1970 (2012).
Meng, X. M., Nikolic-Paterson, D. J. & Lan, H. Y. TGF-β: the master regulator of fibrosis. Nat. Rev. Nephrol. 12, 325–338 (2016).
David, C. J. et al. TGF-β tumor suppression through a lethal EMT. Cell 164, 1015–1030 (2016).
Horiguchi, K. et al. Role of Ras signaling in the induction of Snail by transforming growth factor-β. J. Biol. Chem. 284, 245–253 (2009).
Janda, E. et al. Ras and TGFβ cooperatively regulate epithelial cell plasticity and metastasis: dissection of Ras signaling pathways. J. Cell Biol. 156, 299–314 (2002).
Meno, C. et al. Mouse Lefty2 and zebrafish antivin are feedback inhibitors of Nodal signaling during vertebrate gastrulation. Mol. Cell 4, 287–298 (1999).
Oft, M., Akhurst, R. J. & Balmain, A. Metastasis is driven by sequential elevation of H-ras and Smad2 levels. Nat. Cell Biol. 4, 487–494 (2002).
Sun, X., Meyers, E. N., Lewandoski, M. & Martin, G. R. Targeted disruption of Fgf8 causes failure of cell migration in the gastrulating mouse embryo. Genes Dev. 13, 1834–1846 (1999).
Yamaguchi, T. P., Harpal, K., Henkemeyer, M. & Rossant, J. fgfr-1 is required for embryonic growth and mesodermal patterning during mouse gastrulation. Genes Dev. 8, 3032–3044 (1994).
Zhou, X., Sasaki, H., Lowe, L., Hogan, B. L. & Kuehn, M. R. Nodal is a novel TGF-β-like gene expressed in the mouse node during gastrulation. Nature 361, 543–547 (1993).
Cancer Genome Atlas Research Network. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 32, 185–203 (2017).
Thiagalingam, A. et al. RREB-1, a novel zinc finger protein, is involved in the differentiation response to Ras in human medullary thyroid carcinomas. Mol. Cell. Biol. 16, 5335–5345 (1996).
DaCosta Byfield, S., Major, C., Laping, N. J. & Roberts, A. B. SB-505124 is a selective inhibitor of transforming growth factor-β type I receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 65, 744–752 (2004).
Porsch, H. et al. Efficient TGFβ-induced epithelial–mesenchymal transition depends on hyaluronan synthase HAS2. Oncogene 32, 4355–4365 (2013).
Martin-Malpartida, P. et al. Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors. Nat. Commun. 8, 2070 (2017).
Costello, L. C., Zou, J., Desouki, M. M. & Franklin, R. B. Evidence for changes in RREB-1, ZIP3, and Zinc in the early development of pancreatic adenocarcinoma. J. Gastrointest. Cancer 43, 570–578 (2012).
Kent, O. A., Fox-Talbot, K. & Halushka, M. K. RREB1 repressed miR-143/145 modulates KRAS signaling through downregulation of multiple targets. Oncogene 32, 2576–2585 (2013).
Yamane, T. et al. Transcriptional activation of the cholecystokinin gene by DJ-1 through interaction of DJ-1 with RREB1 and the effect of DJ-1 on the cholecystokinin level in mice. PLoS ONE 8, e78374 (2013).
Kent, O. A. et al. Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes Dev. 24, 2754–2759 (2010).
Winslow, M. M. et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature 473, 101–104 (2011).
Kasai, H., Allen, J. T., Mason, R. M., Kamimura, T. & Zhang, Z. TGF-β1 induces human alveolar epithelial to mesenchymal cell transition (EMT). Respir. Res. 6, 56 (2005).
Schafer, S. et al. IL-11 is a crucial determinant of cardiovascular fibrosis. Nature 552, 110–115 (2017).
Toda, N., Mukoyama, M., Yanagita, M. & Yokoi, H. CTGF in kidney fibrosis and glomerulonephritis. Inflamm. Regen. 38, 14 (2018).
Königshoff, M. et al. WNT1-inducible signaling protein-1 mediates pulmonary fibrosis in mice and is upregulated in humans with idiopathic pulmonary fibrosis. J. Clin. Invest. 119, 772–787 (2009).
Yoshida, S. et al. Extrahepatic platelet-derived growth factor-β, delivered by platelets, promotes activation of hepatic stellate cells and biliary fibrosis in mice. Gastroenterology 147, 1378–1392 (2014).
Dave, N. et al. Functional cooperation between Snail1 and twist in the regulation of ZEB1 expression during epithelial to mesenchymal transition. J. Biol. Chem. 286, 12024–12032 (2011).
Ye, X. et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature 525, 256–260 (2015).
Scheel, C. et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell 145, 926–940 (2011).
Miettinen, P. J., Ebner, R., Lopez, A. R. & Derynck, R. TGF-β induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J. Cell Biol. 127, 2021–2036 (1994).
Shirakihara, T., Saitoh, M. & Miyazono, K. Differential regulation of epithelial and mesenchymal markers by δEF1 proteins in epithelial mesenchymal transition induced by TGF-β. Mol. Biol. Cell 18, 3533–3544 (2007).
Xie, L. et al. Activation of the Erk pathway is required for TGF-β1-induced EMT in vitro. Neoplasia 6, 603–610 (2004).
Peng, G. et al. Spatial transcriptome for the molecular annotation of lineage fates and cell identity in mid-gastrula mouse embryo. Dev. Cell 36, 681–697 (2016).
Wang, Q. et al. The p53 family coordinates Wnt and Nodal inputs in mesendodermal differentiation of embryonic stem cells. Cell Stem Cell 20, 70–86 (2017).
Peiró, S. et al. Snail1 transcriptional repressor binds to its own promoter and controls its expression. Nucleic Acids Res. 34, 2077–2084 (2006).
Lee, J. D., Silva-Gagliardi, N. F., Tepass, U., McGlade, C. J. & Anderson, K. V. The FERM protein Epb4.1l5 is required for organization of the neural plate and for the epithelial–mesenchymal transition at the primitive streak of the mouse embryo. Development 134, 2007–2016 (2007).
Zohn, I. E. et al. p38 and a p38-interacting protein are critical for downregulation of E-cadherin during mouse gastrulation. Cell 125, 957–969 (2006).
Ciruna, B. & Rossant, J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev. Cell 1, 37–49 (2001).
Grande, M. T. et al. Snail1-induced partial epithelial-to-mesenchymal transition drives renal fibrosis in mice and can be targeted to reverse established disease. Nat. Med. 21, 989–997 (2015).
Lovisa, S. et al. Epithelial-to-mesenchymal transition induces cell cycle arrest and parenchymal damage in renal fibrosis. Nat. Med. 21, 998–1009 (2015).
Yoon, S. J., Wills, A. E., Chuong, E., Gupta, R. & Baker, J. C. HEB and E2A function as SMAD/FOXH1 cofactors. Genes Dev. 25, 1654–1661 (2011).
Thuault, S. et al. HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J. Biol. Chem. 283, 33437–33446 (2008).
Acknowledgements
We thank L. Tian and D.-F. Lee for technical assistance, L. Huang for pLenti-HA-Rreb1 plasmid and K. Anderson, A. Nieto and J. P. Thiery for insightful discussions. We acknowledge the support of Y. Furuta and S. Gong of the Mouse Genetics Core, the Molecular Cytology, Integrated Genomics and Flow Cytometry Cores of MSKCC, and S. Y. Kim of the Rodent Genetic Engineering Laboratory of NYU. This work was supported by NIH grants R01CA34610 (J.M.), P01-CA129243 (J.M.), R01DK084391 (A.-K.H.), R01HD094868 (A.-K.H.) and P30-CA008748 (MSKCC). J.S. was supported by an AACR Basic Cancer Research Fellowship (16-40-01-SUJI) and a Charles H. Revson Senior Fellowship in Biomedical Science (17-23). S.M.M. was supported by a Sir Henry Wellcome Postdoctoral Fellowship. Y.-H.H. was supported by a Medical Scientist Training Program grant (T32GM007739) and Predoctoral Fellowship (F30-CA203238) from the National Cancer Institute. H.B. was supported by a Damon Runyon Postdoctoral Fellowship.
Author information
Authors and Affiliations
Contributions
J.S. and J.M. conceived the project and designed the study. J.M., J.S. and A.-K.H. wrote the manuscript. J.S. performed most experiments. S.M.M. and A.-K.H. performed embryo analyses. C.J.D. performed shRNA screens. Q.W. generated E14 Rreb1-knockout clones. E.E.E. and H.B performed molecular cloning of RREB1 phosphorylation-mutant constructs. Y.Z. and Y.-H.H. performed ATAC-seq. W.S. assisted with tissue culture, molecular biology and mouse experiments. J.S., R.C.H. and R.K.S. performed stable isotope labelling with amino acids in cell culture (SILAC) experiments. J.S. and Y.-H.H. performed bioinformatics analyses.
Corresponding author
Ethics declarations
Competing interests
J.M. serves in the scientific advisory board and owns company stock in Scholar Rock.
Additional information
Peer review information Nature thanks Aristidis Moustakas, Angela Nieto and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 RREB1 as a SMAD cofactor in TGF-β gene responses.
a, YFP fluorescence images of CIY organoids expressing KRAS(G12D) under doxycycline control treated with SB505124 or TGF-β for 2.5 days. Scale bars, 200 μm. Images are representative of two independent experiments. b, Influence of KRAS(G12D) on TGF-β gene responses. CIY pancreatic organoids inducibly expressing KRAS(G12D), were treated with SB505124 or TGF-β for 1.5 h and analysed by RNA-seq. Dots represent fold change (in log2) in mRNA levels of individual genes under TGF-β versus SB505124 treatment conditions, with KRAS(G12D) expression turned off (x axis) or on (y axis). Off-diagonal dots correspond to TGF-β gene responses that were enabled (groups I and II) or disabled (groups III and IV) by KRAS(G12D). Gene activation (I and III) and repression responses (II and IV) are included. c, Heat map of four classes of KRAS-modified TGF-β gene responses. n = 1. Representative result of two independent experiments. Classes I–IV correspond to the off-diagonal genes derived from the RNA-seq in b. d, TGF-β gene activation responses augmented by KRAS(G12D) (class I responses) in CIY pancreatic organoids. Fold increase in mRNA levels in TGF-β versus SB505124 treatment conditions in presence or absence of inducible KRAS(G12D). e, Heat maps showing TGF-β induction of Snai1, Has2, Il11, Smad7 and Skil in four independent CIY mouse pancreatic organoid lines with inducible KRAS(G12D) expression. n = 4. f, Heat map of the indicated TGF-β gene responses in spheroid cultures of pancreatic epithelial cells (PECs) inducibly expressing KRAS(G12D). n = 2. g, Heat map of the indicated TGF-β gene responses in monolayer cultures of mouse KrasG12D;Smad4fl/fl;Cdkn2afl/fl;Pdx1-cre (KSIC) PDA cell lines transduced with a SMAD4 vector or an empty vector. n = 2. h, Transcription factor (TF)-binding motifs enriched in KRAS-independent SMAD2/3 binding sites (left) and KRAS-dependent SMAD2/3 binding sites (right). SMAD2/3 ChIP–seq analyses were performed in SMAD4-restored PDA cells that were treated with SB505124 (2.5 μM) or TGF-β (100 pM) for 1.5 h. Transcription factor binding-motif analyses were performed with PscanChIP. n = 821 peak regions (left). n = 778 peak regions (right). i, Motif enrichment analysis of RAS-regulated transcription factors in KRAS-dependent (n = 778 peak regions) and KRAS-independent (n = 821 peak regions) SMAD2/3 binding sites. j, Comparative enrichment of classic SMAD binding motifs (CAGAC and GGCTG) and 5GC motifs (GGC(GC)|(CG)) in a 200-bp region of SMAD2/3 ChIP peaks within 1,000 bp of a transcriptional start site24. The relative enrichment is normalized to the baseline dataset obtained from 20,000 random 200-bp regions from the mm10 genome assembly. The 5GC motifs are enriched approximately fourfold in SMAD2/3 ChIP peaks compared to the baseline, and the classic motifs are enriched twofold.
Extended Data Fig. 2 RREB1 interacts with SMAD and binds to TGF-β target genes.
a, Western blot analysis of RREB1 and HA–RREB1 levels in SMAD4-restored PDA cells stably transduced with a HA–RREB1 vector. Tubulin immunoblotting was used as loading control. Data are representative of two independent experiments. b, Proximity ligation assay showing TGF-β-dependent proximity between RREB1, SMAD2/3 and SMAD4 in the nucleus. Scale bars, 30 μm. Data are representative of two independent experiments. c, Quantification of PLA signals in b. Cell numbers (n) of each group are indicated in the graph, two-tailed unpaired t-test. Data are mean ± s.d. ****P < 0.0001, ***P < 0.001. d, SMAD4-restored PDA cells expressing HA–RREB1 were treated with TGF-β for 1.5 h, lysed and immunoprecipitated (IP) with the indicated antibodies. The immune complexes were collected and subjected to western blot with the antibodies indicated on the left. Data are representative of two independent experiments. e, Heat map of ChIP–seq tag densities for SMAD2/3 and HA–RREB1 in genomic regions ±3 kb from the centre of SMAD2/3 binding peaks in SMAD4-restored PDA cells that were treated with SB505124 or TGF-β for 1.5 h and subjected to SMAD2/3 and HA–RREB1 ChIP–seq analysis. ChIP–seq was performed once, and an independent ChIP was performed in which selective genomic regions were confirmed by qPCR.
Extended Data Fig. 3 RREB1 is phosphorylated and regulated by ERK.
a, Representative immunofluorescence images of HA–RREB1 in SMAD4-restored PDA cells treated with DMSO or 1 μM ERK inhibitor SCH772984 (ERKi) for 6 h. Scale bar, 20 μm. Data are representative of two independent experiments. b, c, Western blot analysis of RREB1 (b) or HA–RREB1 levels (c) in SMAD4-restored PDA cells treated with DMSO, 1 μM ERKi or 1 μM AZD6244 (MEKi; an inhibitor of the ERK-activating kinases MEK1/2) for the indicated time periods. Tubulin immunoblotting was used as loading control. Data are representative of two independent experiments. d, ChIP–PCR analysis of HA–RREB1 binding to the indicated sites (Figs. 1g, 3b) in Snai1, Has2 and Il11 in SMAD4-restored PDA cells that were treated with vehicle (DMSO) or ERKi (1 μM) for 6 h. Mean ± s.e.m. n = 3, two-way ANOVA. **P < 0.01, ***P < 0.001, ****P < 0.0001. e, SMAD4-restored PDA cells expressing HA–RREB1 were treated with ERKi for the indicated length of time. HA–REBB1 was tested for binding to Snai1 DE2 and Has2 PP1 double-stranded DNA oligonucleotide probes in DNA affinity precipitation assays. Data are representative of two independent experiments. f, Schematic of RREB1. Each tick represents a previously annotated phosphorylation site in PhosphoSitePlus identified in at least two independent mass spectrometry experiments. Red filled circles represent high stoichiometry (>15%) phosphorylation sites that are inhibited by ERKi, as identified in g. Zinc-finger domains annotated in Uniprot are shown. g, Phosphorylation stoichiometry of four ERK-dependent RREB1 phosphorylation sites in SMAD4-restored PDA cells, as determined by SILAC mass spectrometry of cells treated with DMSO (control) in light medium or ERKi in heavy medium for 6 h. h, Summary of ERK-dependent RREB1 phosphorylation sites, sequence motifs and phosphorylation stoichiometry. i, Rreb1-KO PDA cells were transduced with the indicated RREB1-WT or phosphorylation-site mutant constructs, then treated with SB505124 or TGF-β for 1.5 h. mRNA levels of Snai1 and Has2 were determined by qPCR with reverse transcription. Mean ± s.e.m. n = 4, two-way ANOVA. ***P < 0.001, ****P < 0.0001. j, ChIP–PCR analysis of HA–RREB1 binding to the indicated sites in Rreb1-KO PDA cells transduced with the indicated RREB1-WT or phosphorylation-site mutant constructs. Mean ± s.e.m. n = 4, two-tailed unpaired t-test. ****P < 0.0001.
Extended Data Fig. 4 RREB1 mediates KRAS-dependent TGF-β responses in PDA cells.
a, Scheme of CRISPR–Cas9-mediated mutation of Rreb1 in mouse SMAD4-restored PDA cells. b, sgRNA sequences and genomic sequences of Rreb1 coding region (CDS) exons 1 and 7 in mutant clones KO1 and KO2 derived from SMAD4-restored PDA cells. c, Western blot analysis of RREB1 levels in WT and Rreb1-KO cells. Tubulin immunoblotting was used as loading control. Data are representative of two independent experiments. d, Western blot analysis of E-cadherin in mouse KSIC PDA cells, SMAD4-restored PDA cells and two Rreb1-KO SMAD4-restored PDA clones, treated with SB505124 or TGF-β for 24 h. Tubulin immunoblotting was used as loading control. Data are representative of two independent experiments. e, Representative E-cadherin immunofluorescence and DAPI staining of the same cells as in d treated with SB505124 or TGF-β for 48 h. Scale bars, 100 μm. Data are representative of two independent experiments. f, Gene track view of SMAD2/3 ChIP–seq tags in the Smad7 locus of the WT and Rreb1-KO PDA cells. The gene body is schematically represented at the bottom. ChIP–seq was performed once and an independent ChIP was performed in which selective genomic regions were confirmed by qPCR. g, mRNA levels of Snai1, Has2 and Il11 in WT and two Rreb1-KO cells that were transduced with an RREB1 vector or empty vector and then treated with SB505124 or TGF-β for 1.5 h. Mean ± s.e.m. n = 4; two-way ANOVA; ****P < 0.0001.
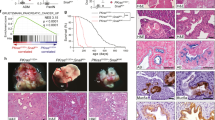
Extended Data Fig. 5 RREB1 mediates tumorigenic EMT in lung adenocarcinoma cells.
a, Snai1, Has2 and Il11 mRNA levels in 393T3 mouse LUAD cells treated with DMSO (Ctrl) or ERKi (SCH772984, 1 μM) for 6 h, followed by treatment of SB505124 or TGF-β for 1.5 h. Mean ± s.e.m. n = 4; two-way ANOVA; ****P < 0.0001. b, Cdh1 mRNA levels in 393T3 cells with the indicated treatments for 48 h. Mean ± s.d. n = 4; two-way ANOVA. c, Western blot analysis of E-cadherin in 393T3 cells with the indicated treatments for 48 h. Tubulin immunoblotting was used as loading control. Data are representative of two independent experiments. d, SMAD4-restored PDA cells and 393T3 LUAD cells cultured in D10F containing 2.5 μM MK220612 were treated with SB505124 (2.5 μM) or TGF-β (100 pM) and assayed for cleaved caspase 3/7 activity at the indicated times. Mean ± s.e.m. n = 4; two-way ANOVA; ***P < 0.001. e, SMAD4-restored PDA cells and 393T3 cells cultured in D10F containing 2.5 μM MK2206 were treated with SB505124 or TGF-β. Cell viability was determined at the indicated times. Mean ± s.e.m. n = 4; two-way ANOVA; ***P < 0.001. f, sgRNA sequence targeting Rreb1 CDS exon 3, and mutant Rreb1 genomic sequences of the resulting 393T3 KO1 and KO2 clones. g, mRNA levels of Snai1 and Has2 in the WT and Rreb1-KO 393T3 cells after treatment with SB505124 (2.5 μM) or TGF-β (100 pM) for 1.5 h. Mean ± s.e.m. n = 4; two-way ANOVA; ****P < 0.0001. h, Phase contrast images of 393T3 cell monolayers treated with SB505124 or TGF-β for 48 h. Scale bars, 200 μm. Data are representative of two independent experiments. i, Weight and volume of tumours in Fig. 2i. Mean ± s.e.m. n = 10, two sites were inoculated per mouse; two-tailed unpaired t-test; ****P < 0.0001. j, Representative haematoxylin and eosin staining images of indicated lung tissue sections from Fig. 2j. Scale bars, 200 μm. Data are representative of two independent experiments.
Extended Data Fig. 6 RREB1-dependent TGF-β responses in LUAD and PDA cells.
a, sgRNA sequence targeting RREB1 CDS exon 3 and mutant RREB1 genomic sequences of the resulting A549 KO1 and KO2 clones. b, SNAIL and SLUG mRNA levels in WT A549 and two RREB1 KO clones treated with SB505124 or TGF-β for 24 h. Mean ± s.e.m. n = 4; two-way ANOVA; ****P < 0.0001. c, Phase-contrast images of WT A549 and RREB1-KO cell monolayers treated with SB505124 or TGF-β for 48 h. Scale bars, 200 μm. Data are representative of two independent experiments. d, Growth kinetics of tumours formed by subcutaneously inoculated WT or RREB1-KO A549 cells in athymic mice, as determined by BLI of a transduced firefly luciferase gene in the cells. Mean ± s.e.m. n = 10, two sites were inoculated per mouse; two-way ANOVA. e, Gene ontology analysis of TGF-β response genes in CIY organoids inducibly expressing KRAS(G12D), based on the RNA-seq in Extended Data Fig. 1b. f, WT and Rreb1-KO PDA cells were treated with SB505124 or TGF-β for 1.5 h and analysed by RNA-seq. Dots represent fold change (in log2) in mRNA levels of individual genes under TGF-β versus SB505124 treatment conditions, in Rreb1-KO (x axis) or WT cells (y axis). Off-diagonal dots corresponding to Snai1, Has2, Il11 and Wisp1 are highlighted. g, Induction of Snai1 and Zeb1 expression by TGF-β in mouse PDA cells. Mean ± s.d. n = 4. h, sgRNA sequence targeting Snai1 and resulting mutant Snai1 genomic sequences in mouse PDA cells. i, Knockdown of Zeb1 with two independent shRNAs in SNAIL-KO mouse PDA cells (KOsh cells). Mean ± s.d. n = 4. j, Fibrogentic gene responses to TGF-β in WT and SNAIL and ZEB1-double depleted KOsh PDA cells. Mean ± s.d. n = 4. k–m, E-cadherin levels (k), phase-contrast images (l) and E-cadherin and Zeb1 immunofluorescence in WT and KOsh PDA cells that were treated with SB505124 or TGF-β for 48 h. Scale bars, 100 μm. Data are representative of two independent experiments.
Extended Data Fig. 7 RREB1-dependent TGF-β responses in mammary epithelial cells.
a, sgRNA sequence targeting RREB1 CDS exon 3 and mutant RREB1 genomic sequences of the resulting NMuMG KO1 and KO2 clones. b, Phase-contrast images of WT and Rreb1-KO NMuMG cell monolayers treated with SB505124 or TGF-β for 48 h. Scale bar, 100 μm. Data are representative of two independent experiments. c, Western blot analysis of E-cadherin in WT and Rreb1-KO NMuMG cells, treated with SB505124 or TGF-β for 48 h. β-actin immunoblotting was used as loading control. Data are representative of two independent experiments. d, WT and Rreb1-KO NMuMG cells were treated with SB505124 or TGF-β for 1.5 h and analysed by RNA-seq. Dots represent fold change (in log2) in mRNA levels of individual genes under TGF-β versus SB505124 treatment conditions, in Rreb1-KO (x axis) or WT cells (y axis). Off-diagonal dots corresponding to Snai1 and Has2 are highlighted. e, ChIP–PCR analysis of SMAD2/3 binding to the Snai1 (DE2) and Has2 (UE1) regions (Fig. 1g) in WT and Rreb1-KO NMuMG cells. Cells were treated with 2.5 μM SB505125 or 100 pM TGF-β for 1.5 h. Mean ± s.e.m. n = 4; two-way ANOVA; ***P < 0.001, ****P < 0.0001. f, mRNA levels of Snai1 and Has2 in WT and Rreb1-KO NMuMG cells after treatment with SB505124 or TGF-β for 1.5 h. Mean ± s.e.m. n = 4; two-way ANOVA; ****P < 0.0001. g, ChIP–PCR analysis of HA–RREB1 binding to the indicated Snai1 and Has2 regions in NMuMG cells that were treated with vehicle (DMSO) or the ERKi SCH772984 (1 μM) for 6 h. Mean ± s.e.m. n = 3; two-tailed unpaired t-test. h, Snai1 and Has2 mRNA levels in NMuMG cells treated with DMSO (Ctrl), ERKi (1 μM SCH772984), EGF (10 ng ml−1, 10 min) or EGFR inhibitor (gefitinib, 1 μM, 2 h), followed by SB505124 or TGF-β treatment for another 1.5 h. Mean ± s.e.m. n = 4; two-way ANOVA; ***P < 0.001, ****P < 0.0001.
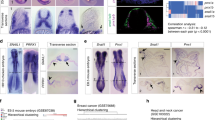
Extended Data Fig. 8 RREB1 in gastrulation EMT and mesendoderm differentiation.
a, Corn plot presentation of Rreb1, Snai1, Cdh2, Gsc and T in E7.0 mouse embryo. A, anterior; L, left; R, right; P, posterior regions. Each dot represents transcript level at a specific positional address. Heat map denotes expression level of each gene computed from transcript counts in RNA-seq datasets41. b, Reads per million reads (RPM) of Rreb1, Snai1, Twist1, Cdh2, Eomes and Zeb2 in the RNA-seq dataset at the indicated times after shifting ES cells into LIF-deficient embryoid body differentiation medium. c, sgRNA sequence targeting Rreb1 CDS exon 3, and mutant Rreb1 genomic sequences of four resulting mouse ES cell KO clones. d, mRNA levels of EMT (Snai1 and Cdh2) and mesendoderm differentiation genes (Eomes, Gsc, T and Mixl1) in WT and four independent Rreb1-KO clones on day 4 embryoid body differentiation. Mean ± s.d. n = 4; two-way ANOVA; ****P < 0.0001. e, mRNA levels of the indicated genes in WT and four independent Rreb1-KO clones treated with receptor inhibitor (SB505125) or activin A (AC) for 2 h. Mean ± s.e.m. n = 4; two-way ANOVA; ****P < 0.0001. f, Gene set enrichment analysis for gastrulation, EMT and stem cell differentiation genes in WT cells, and absence in Rreb1-KO cells at day 4 embryoid body differentiation.
Extended Data Fig. 9 RREB1 and SMAD contextually regulate EMT genes.
a, Heat map of ChIP–seq tag densities for SMAD2/3 and HA-RREB1 in genomic regions ±3 kb from centre of 3,422 high-confidence SMAD2/3 binding peaks in day 3 embryoid bodies subjected to SMAD2/3 and HA ChIP–seq analyses. b, Gene track view of SMAD2/3 and HA-RREB1 ChIP–seq tags in the loci of EMT genes (Has2, Twist1 and Zeb1) and early mesendoderm lineage genes (Eomes, T and Mixl1) in day 3 embryoid bodies. Gene bodies are schematically represented at the bottom of each track set. c, Gene track view of ATAC-seq and SMAD2/3 and RREB1 ChIP–seq tags on indicated loci, in day 3 embryoid bodies (red tracks) versus TGF-β treated (1.5 h) SMAD4-restored PDA cells (blue tracks). In a–c, ChIP–seq was performed once and an independent ChIP was performed in which selective genomic regions were confirmed by qPCR.
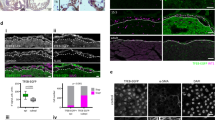
Extended Data Fig. 10 Rreb1−/− mouse embryo chimaeras exhibit defects in early development.
a, E7.5 and E8.5 chimeric embryos containing WT ES cells or Rreb1−/− ES cells were scored, on the basis of gross morphology, as normal or mild defects, developmentally retarded or severely abnormal. At E7.5, a fraction of Rreb1+/+ ES cell embryos displayed small clumps of cells in the amniotic cavity, possibly an artefact from the microinjection, and were therefore scored as abnormal. Rreb1−/− data are compiled from four distinct KO clones. b, Bright-field morphology and mCherry fluorescence (marking descendants of injected ES cells) in representative litters of Rreb1−/− ES-cell-containing chimeric embryos dissected at E7.5 and E8.5. nc, non-chimeric; lc, low chimaerism. Asterisks mark morphologically abnormal or developmentally retarded embryos. c, Bright-field images of morphologically abnormal Rreb1−/− ES-cell-containing chimeric E8.5 embryos. Embryos exhibited abnormal headfold development, including disproportionate headfolds (i) and asymmetric headfolds (ii). Axis duplication was also observed, (iii) and (iv). Of note, the embryo in (iii) is also developmentally retarded. d, e, Confocal maximum intensity projections of whole-mount immunostained E8.5 Rreb1−/− ES-cell-containing chimeric embryos. d, An embryo with an ectopic somite-like structure (arrowhead). e, The embryo in c (iv) with axis duplication of the headfolds. f, Sagittal confocal optical sections of whole-mount immunostained chimeric E7.5 embryos. Embryos shown in f (i, ii) have multiple cavities and multiple expression sites of SNAIL, hence anterior–posterior axis orientation is not possible. g, Bright-field images of morphologically abnormal Rreb1−/− ES-cell-containing chimeric E7.5 embryos. Embryos frequently had protrusions into the cavity and thickening of the posterior epiblast, marked by arrowheads. h, i, Confocal maximum intensity projections of chimeric embryos after whole-mount immunostaining for phospho-histone H3 (h), labelling mitotic cells, and cleaved caspase 3 (i), labelling apoptotic cells. Brackets demarcate the primitive streak. j, Sagittal confocal optical sections of chimeric E7.5 embryos after whole-mount immunostaining for E-cadherin and N-cadherin. Arrowhead, aberrant N-cadherin expression. Scale bars, 50 µm. Images in b–j are representative of two independent experiments.
Supplementary information
Supplementary Information
This file contains Supplementary Methods.
Supplementary Figures
Supplementary Figure 1: Western blot gel source data.
Video 1
: KRAS organoids undergo lethal EMT with TGF-β treatment. CIY pancreatic organoids with KRASG12D off (left panel) or KRASG12D on (right panel) were treated with TGF-β, and then monitored by time-lapse imaging for 52 h.
Rights and permissions
About this article
Cite this article
Su, J., Morgani, S.M., David, C.J. et al. TGF-β orchestrates fibrogenic and developmental EMTs via the RAS effector RREB1. Nature 577, 566–571 (2020). https://doi.org/10.1038/s41586-019-1897-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1897-5
This article is cited by
-
Biomaterial-based mechanical regulation facilitates scarless wound healing with functional skin appendage regeneration
Military Medical Research (2024)
-
Decoding gene regulatory circuitry underlying TNBC chemoresistance reveals biomarkers for therapy response and therapeutic targets
npj Precision Oncology (2024)
-
A prognostic marker LTBP1 is associated with epithelial mesenchymal transition and can promote the progression of gastric cancer
Functional & Integrative Genomics (2024)
-
A spatiotemporal atlas of mouse liver homeostasis and regeneration
Nature Genetics (2024)
-
Elucidating shared biomarkers and pathways in kidney stones and diabetes: insights into novel therapeutic targets and the role of resveratrol
Journal of Translational Medicine (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.