Abstract
Mammals form scars to quickly seal wounds and ensure survival by an incompletely understood mechanism1,2,3,4,5. Here we show that skin scars originate from prefabricated matrix in the subcutaneous fascia. Fate mapping and live imaging revealed that fascia fibroblasts rise to the skin surface after wounding, dragging their surrounding extracellular jelly-like matrix, including embedded blood vessels, macrophages and peripheral nerves, to form the provisional matrix. Genetic ablation of fascia fibroblasts prevented matrix from homing into wounds and resulted in defective scars, whereas placing an impermeable film beneath the skin—preventing fascia fibroblasts from migrating upwards—led to chronic open wounds. Thus, fascia contains a specialized prefabricated kit of sentry fibroblasts, embedded within a movable sealant, that preassemble together diverse cell types and matrix components needed to heal wounds. Our findings suggest that chronic and excessive skin wounds may be attributed to the mobility of the fascia matrix.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Marshall, C. D. et al. Cutaneous scarring: basic science, current treatments, and future directions. Adv. Wound Care 7, 29–45 (2018).
Finnerty, C. C. et al. Hypertrophic scarring: the greatest unmet challenge after burn injury. Lancet 388, 1427–1436 (2016).
Morton, L. M. & Phillips, T. J. Wound healing and treating wounds: differential diagnosis and evaluation of chronic wounds. J. Am. Acad. Dermatol. 74, 589–605, quiz 605–606 (2016).
Do, N. N. & Eming, S. A. Skin fibrosis: models and mechanisms. Curr. Res. Transl. Med. 64, 185–193 (2016).
Sen, C. K. et al. Human skin wounds: a major and snowballing threat to public health and the economy. Wound Repair Regen. 17, 763–771 (2009).
Hinz, B. Myofibroblasts. Exp. Eye Res. 142, 56–70 (2016).
Driskell, R. R. et al. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature 504, 277–281 (2013).
Greenhalgh, S. N., Conroy, K. P. & Henderson, N. C. Healing scars: targeting pericytes to treat fibrosis. QJM 108, 3–7 (2015).
Plikus, M. V. et al. Regeneration of fat cells from myofibroblasts during wound healing. Science 355, 748–752 (2017).
Shook, B. A. et al. Myofibroblast proliferation and heterogeneity are supported by macrophages during skin repair. Science 362, eaar2971 (2018).
Mori, L., Bellini, A., Stacey, M. A., Schmidt, M. & Mattoli, S. Fibrocytes contribute to the myofibroblast population in wounded skin and originate from the bone marrow. Exp. Cell Res. 304, 81–90 (2005).
Rinkevich, Y. et al. Identification and isolation of a dermal lineage with intrinsic fibrogenic potential. Science 348, aaa2151 (2015).
Jiang, D. et al. Two succeeding fibroblastic lineages drive dermal development and the transition from regeneration to scarring. Nat. Cell Biol. 20, 422–431 (2018).
Adstrum, S., Hedley, G., Schleip, R., Stecco, C. & Yucesoy, C. A. Defining the fascial system. J. Bodyw. Mov. Ther. 21, 173–177 (2017).
Stecco, C. & Schleip, R. A fascia and the fascial system. J. Bodyw. Mov. Ther. 20, 139–140 (2016).
Dunkin, C. S. et al. Scarring occurs at a critical depth of skin injury: precise measurement in a graduated dermal scratch in human volunteers. Plast. Reconstr. Surg. 119, 1722–1734 (2007).
Koehler, R. H. et al. Minimal adhesions to ePTFE mesh after laparoscopic ventral incisional hernia repair: reoperative findings in 65 cases. JSLS 7, 335–340 (2003).
Rippa, A. L., Kalabusheva, E. P., & Vorotelyak, E., A. Regeneration of dermis: scarring and cells involved. Cells 8, 607 (2019).
Zamir, E. A., Rongish, B. J. & Little, C. D. The ECM moves during primitive streak formation—computation of ECM versus cellular motion. PLoS Biol. 6, e247 (2008).
Szabó, A., Rupp, P. A., Rongish, B. J., Little, C. D. & Czirók, A. Extracellular matrix fluctuations during early embryogenesis. Phys. Biol. 8, 045006 (2011).
Aleksandrova, A. et al. Convective tissue movements play a major role in avian endocardial morphogenesis. Dev. Biol. 363, 348–361 (2012).
Loganathan, R. et al. Extracellular matrix motion and early morphogenesis. Development 143, 2056–2065 (2016).
Miron-Mendoza, M., Koppaka, V., Zhou, C. & Petroll, W. M. Techniques for assessing 3-D cell-matrix mechanical interactions in vitro and in vivo. Exp. Cell Res. 319, 2470–2480 (2013).
Sakar, M. S. et al. Cellular forces and matrix assembly coordinate fibrous tissue repair. Nat. Commun. 7, 11036 (2016).
Abu-Hijleh, M. F., Roshier, A. L., Al-Shboul, Q., Dharap, A. S. & Harris, P. F. The membranous layer of superficial fascia: evidence for its widespread distribution in the body. Surg. Radiol. Anat. 28, 606–619 (2006).
Avelar, J. Regional distribution and behavior of the subcutaneous tissue concerning selection and indication for liposuction. Aesthetic Plast. Surg. 13, 155–165 (1989).
Lockwood, T. E. Superficial fascial system (SFS) of the trunk and extremities: a new concept. Plast. Reconstr. Surg. 87, 1009–1018 (1991).
Acknowledgements
We thank G. Mettenleiter for technical assistance with the SEM; S. Dietzel and the Core Facility Bioimaging at the Biomedical Centre of the Ludwig-Maximilians-Universität München for access and support with the multi-photon system. Y.R. was supported by the Human Frontier Science Program Career Development Award (CDA00017/2016), the German Research Foundation (RI 2787/1-1 AOBJ: 628819), the Fritz–Thyssen–Stiftung (2016-01277) and the European Research Council Consolidator Grant (ERC-CoG 819933). D.C.-G. was supported by the Consejo Nacional de Ciencia y Tecnología (CONACYT) and the Deutscher Akademischer Austauschdienst (DAAD).
Author information
Authors and Affiliations
Contributions
Y.R. outlined and supervised the research narrative and experimental design. D.C.-G. performed the cell and matrix tracing experiments, chimeric skin transplantations, ePTFE implantations and image analysis. D.J. performed the cell depletion with DT in pups, flow cytometry experiments and analysis, fascia release experiments and experiments with human skin samples. S.C. performed the live imaging and analysis. S.C. and P.R. performed 3D imaging and analysis. H.Y. performed histology and immunofluorescence staining of human samples. J.W. provided veterinary advice and prepared animal experiment protocols. S.K.G. made the viral particles and performed titre quantification. Q.Y. assisted in the flow cytometry experiments. U.M. and T.V. collected consent from patients and the primary human tissue samples, and assisted with translational and clinical advice. U.M. wrote, in part, the ethical application for collection and use of human samples. M.A. and A.W. performed the scanning electron microscopy images. Y.R., D.C.-G. and D.J. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peer review information Nature thanks Christopher Buckley and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Fig. 1 Fate mapping of fascia cells with DiI.
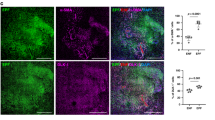
a, DiI labelling of fascia cells. b, Histology showing DiI+ cells in uninjured controls (left) and at 14 dpw (right). Representative images of five biological replicates. c, Immunolabelling (top) and fractions (bottom) of DiI-positive cells expressing mesenchymal/fibroblast markers ITGB1, ER-TR7, THY1 and PDGFRα. Data are mean ± s.e.m.; n = 4 (5 in ITGB1) images analysed from 3 biological replicates. d, Immunolabelling (top) and fractions (bottom) of DiI-positive monocytes/macrophages (MOMA-2), lymphatic cells (LYVE1), endothelial cells (PECAM1) and nerve cells (TUBB3). Data are mean ± s.e.m.; n = 5 images analysed from 3 biological replicates. Lines delimit PC. The dotted line delimits the wound. Scale bars, 200 μm.
Extended Data Fig. 2 Fascia EPFs traverse PC.
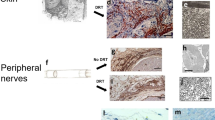
a, Gating strategy for fibroblasts analysis. b, Percentages of fibroblasts (Lin−) and lineage-positive cells in fascia and dermis. Data are mean ± s.e.m.; n = 4 independent experiments. Unpaired two-tailed t-test, 95% CI. c, Scatter plots of EPFs (GFP+Lin−) and ENFs (TdTomato+Lin−) in fascia and dermis. Representative plots of three independent experiments. d, EPF and ENF fractions in fascia and dermis. Data are mean ± s.e.m.; n = 4 independent experiments. Two-way ANOVA, multiple comparison Tukey’s test, 95% CI. e, Endothelial cell (PECAM1+), lymphatic cell (LYVE1+), macrophages (F4/80+) and nerve cell (NGFR+) fractions in fascia and dermis. Data are mean ± s.e.m.; n = 3 biological replicates. Two-way ANOVA, multiple comparison Tukey’s test, 95% CI. f–k, Representative images of 3D-rendered En1cre;R26mTmG or En1cre;R26VT2/Gk3 back skin fascia from at least three biological replicates. f, Lateral view (left) and cross-sections (right) of adult fascia. g, Top view (ventral side up) of neonate back skin. h, Top side view (top) and lateral cross-section (bottom) at the forelimb junction showing EPF traversing the PC. i, Top view at a muscle breach showing EPFs in both locations. j, Top view at a muscle opening where nerves pass through and polyclones of EPFs reside. k, Top view (top, epidermis side up) and lateral cross-section (bottom) of an adult superficial wound (3 dpw). Brocken lines delimit PC. Dotted lines delimit the epidermis. Scale bars, 1,500 μm (g), 100 μm (f, i, j) and 500 μm (k); v, vessels; nb, nerve bundles.
Extended Data Fig. 3 Fascia EPFs maintain position in steady conditions and recede from wounds over time.
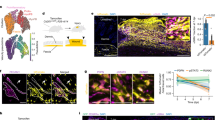
a, Dermal versus fascia EPFs chimeras in uninjured conditions. b, Fascia (left) or dermal (right) EPFs-traced chimeras. Representative images of 3 biological replicates. c, Scars at 70 dpw from deep injuries of fascia EPF-traced chimeras immunolabelled for DPP4. Representative images of 3 biological replicates. d, Cleaved CASP3 expression in wounds from fascia (left) or dermal (right) EPF-traced chimeras from deep (top) or superficial (bottom) injuries at 14 dpw. e, Fractions of fascia or dermal EPFs in the wound, dermis or fascia control regions positive for cleaved CASP3. Data are mean ± s.e.m.; n = 6 and 5 (fascia and dermal EPF, respectively) images analysed from 5 biological replicates. Lines delimit the border between fascia and dermis. Dotted lines delimit the wound or scar. Scale bars, 200 μm.
Extended Data Fig. 4 Fascia EPFs express wound fibroblast markers.
a, Dermal versus fascia EPF-traced chimeras with two injury conditions. b–f, Representative immunolabelling for the fibroblast markers DPP4 (b), DLK1 (c), CD24 (d), CD34 (e) and LY6A (f) from 4 biological replicates. g, Areas analysed (top) for marker-positive EPF quantification (bottom). Data are mean ± s.e.m.; n = 4 (DPP4 in dermal EPFs), 5 (all markers in dermal EPFs with the exception of DPP4 and all markers in fascia EPFs with the exception of DLK1, CD34 and LY6A), 6 (DLK1 in fascia EPFs), 7 (CD34 in fascia EPFs) or 11 (LY6A in fascia EPFs) images analysed from 4 biological replicates. One-way ANOVA, multiple comparison Tukey’s test, 95% CI. Dotted lines delimit the wound bed. Scale bars, 200 μm.
Extended Data Fig. 5 Differential expression of classical markers on fascia and dermal fibroblasts.
a, Gating strategy for fibroblast (Lin−) cytometry. b, Histogram plots of fibroblast-marker expression in fascia or dermis derived fibroblasts from three biological replicates. c, Fraction of marker-positive cells from total fibroblast population. Data are mean ± s.e.m.; n = 3 biological replicates. Unpaired two-tailed t-test, 95% CI. d, Gating strategy for fascia EPF (Lin− GFP+) sorting and detection of LY6A, PDGFR1, DPP4 and ITGB1 expression. Representative plots of three biological replicates.
Extended Data Fig. 6 Fascia but not dermal matrix steers into wounds.
a, Matrix tracing in chimeric grafts. b, Grafts at 7 dpw immunolabelled for collagen I, III and VI. Representative image of three biological replicates. c, Label coverage fraction from total collagens in the wound at defined time points. Data are mean ± s.e.m.; n = 4 (7 dpw) and 9 (14 dpw) sections analysed from 3 biological replicates. Unpaired two-tailed t-test, 95% CI. d, Wounds at 14 dpw immunolabelled for collagens. Representative images of three biological replicates. e–f, Higher magnification of the insets in d. g, h, Double matrix tracing in deep-injured dermal EPF-traced grafts at 7 dpw. Representative image of three biological replicates. i, j, Double matrix tracing in deep-injured fascia EPF-traced grafts at 14 dpw. Representative image of three biological replicates. k, l, Double matrix labelling in superficial-injured dermal EPF-traced grafts at 14 dpw immunolabelled for collagens. Representative image of three biological replicates. Dotted lines delimit the wound. Arrowheads mark the original injury. Continuous lines delimit the epidermis dermis margin. Scale bars, 500 μm (b), 100 μm (d–f) and 200 μm (h, j, l).
Extended Data Fig. 7 Fascia matrix forms the eschar and is remodelled in the wound.
a, Left, in situ matrix tracing and collagen I, III and VI immunolabelling at defined time points after wounding. Representative images of three biological replicates. Right, subsampled fractal dimension maps of the FITC signal of uninjured tissue and at 3 and 7 dpw, and the collagen signal at 14 dpw. b, Matrix label coverage from total collagen I, III and VI signal in the wound. Data are mean ± s.e.m.; n = 3 (uninjured), 4 (3 dpw), 7 (7 dpw) and 4 (14 dpw) sections analysed from 3 biological replicates. One-way ANOVA, Tukey’s multiple comparisons. c, d, Average fractal dimension (c) and lacunarity (d) from subsampled maps. Data are mean ± s.e.m.; n = 5 (uninjured), 5 (3 dpw), 8 (7 dpw) and 3 (14 dpw) images analysed from 3 biological replicates. One-way ANOVA, Tukey’s test, 95% CI. e, Scatter plot of average fractal dimension and lacunarity values. f, In situ matrix tracing and SELP immunolabelling at defined time points after wounding. Representative images of three biological replicates. The broken line separates dermis from fascia. Dotted lines indicate the wound. Scale bars, 200 μm.
Extended Data Fig. 8 Inflammation resolution and coagulation stay unaffected during fascia blocking.
a, Sham (left) or ePTFE-implanted (right) wounds at 7 (top) or 63 dpw (bottom) immunolabelled for collagen I, III and VI. Representative images of three biological replicates. b–i, Immunolabellings (left) and fractions (right) of immune cells (PTPRC+; b), neutrophils (LY6G+; c), T cells (CD3+; d), NK cells (NCR1+; e), B cells (CD19+; f), macrophages and monocytes (MOMA2+; g) and cells expressing the pro- and anti-inflammatory cytokines (IL12 (h) and IL4 (i)). Mean with SEM, n = 3 images analysed from 3 biological replicates. Unpaired two-tailed t-test, 95% CI (b). One-way ANOVA, Tukey’s test, 95% CI (c-i). j, Activated platelets (SELP) in 7 dpw sham and ePTFE-implanted wounds. Representative images of 3 biological replicates. k, Mean grey value of SELP signal. Data are mean ± s.e.m.; n = 3 images analysed from 3 biological replicates. Two-tailed Student’s t-test, 95% CI. Dotted lines delimit the wound area. Scale bars, 200 μm (main images) and 100 μm (magnified insets).
Extended Data Fig. 9 EPFs steer matrix in vitro independently of proliferation.
a, En1cre;R26iDTR biopsies at day 0 and 6 after short treatment with DT or vehicle, immunolabelled for collagen I, III and VI. Representative images of three replicates. b, Collagens density. Data are mean ± s.e.m.; n = 3 images analysed from 3 biological replicates. Two-way ANOVA, multiple comparison Tukey’s test, 95% CI. c, Cell density. Data are mean ± s.e.m.; n = 3 images analysed from 3 biological replicates. One-way ANOVA, multiple comparison Tukey’s test, 95% CI. d, e, Time-lapse images (d) and contraction rate (e) of En1cre;R26iDTR neonate fascia biopsy in culture treated with DT for 1 h. Representative samples from three replicates. Contraction values obtained from Supplementary Videos 4, 6. f, Fascia biopsies treated with etoposide and immunolabelled for MKI67. g, Fraction of MKI67+ cells. Data are mean ± s.e.m.; n = 3 images analysed from 3 biological replicates. One-way ANOVA, Dunnett’s multiple comparisons, 95% CI. h, i, Time-lapse images (h) and contraction rate (i) of neonate fascia biopsy in culture treated with 100 μM etoposide. Representative samples from three replicates. Contraction values obtained from Supplementary Videos 4, 7. j, Mean (± s.e.m.) matrix contraction velocity during the first 25 h of imaging; n = 25 values from Supplementary Video 7. Two-tailed Student’s t-test, 95% CI. Lines show the distance between two tracked points in the SHG channel. Scale bars, 50 μm (f), 200 μm (a, h) and 500 μm (d).
Extended Data Fig. 10 Fascia matrix steering precedes proliferation in vivo.
a, In situ fascia matrix labelling and EdU pulses. b, EdU detection in sections at defined time points. c, Fraction of EdU+ cells in the wound from total EdU+ cells. Data are mean ± s.e.m.; n = 3 (uninjured), 4 (3 dpw), 6 (7 dpw) and 4 (14 dpw) images analysed from 3 biological replicates. One-way ANOVA, Tukey’s multiple comparisons. Arrows indicate EdU-positive nuclei. Arrowheads indicate the original injury site. Broken and dotted lines delimit fascia and wounds respectively. Scale bars, 200 μm.
Supplementary information
Video 1
: 3D reconstruction of adult En1Cre;R26mTmG fascia. EPFs in green and SHG in cyan
Video 2
: 3D reconstruction of neonate En1Cre;R26mTmG back-skin and fascia. EPFs in green and TdTomato in red. Anterior-to-posterior axis from top to bottom of the video frame. Mid-to-lateral axis from left to right of the video frame
Video 3
: 3D reconstruction of adult En1Cre;R26mTmG back-skin wound 3 dpw. EPFs in green, TdTomato in red, SHG in cyan, and collagen I in magenta
Video 4
: Time-lapse of 3D rendered P0 C57BL6/J fascia biopsy in culture. Second harmonic generation (SHG) in cyan and autofluorescence in green
Video 5
: 3D reconstruction of day 14 wounds after transplantation of chimeric grafts with labeled matrix. TdTomato (Skin) in red, GFP (Fascia) in green, NHS AF647 in magenta, and collagen I+III+VI immunolabeling in blue. Skin to fascia orientation from top to bottom of the video frame
Video 6
: Time-lapse of 3D rendered P0 En1Cre;R26iDTR fascia biopsy in culture treated with an acute exposure of 2 μg/ml of diphtheria toxin. Second harmonic generation (SHG) in cyan and autofluorescence in green
Video 7
: Time-lapse of 3D rendered P0 C57BL6/J fascia biopsy in culture treated with 100 μM etoposide. Second harmonic generation (SHG) in cyan and autofluorescence in green
Source data
Rights and permissions
About this article
Cite this article
Correa-Gallegos, D., Jiang, D., Christ, S. et al. Patch repair of deep wounds by mobilized fascia. Nature 576, 287–292 (2019). https://doi.org/10.1038/s41586-019-1794-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1794-y
This article is cited by
-
Fractal analysis of extracellular matrix for observer-independent quantification of intestinal fibrosis in Crohn’s disease
Scientific Reports (2024)
-
Local and systemic mechanisms that control the hair follicle stem cell niche
Nature Reviews Molecular Cell Biology (2024)
-
Cellular and molecular mechanisms of skin wound healing
Nature Reviews Molecular Cell Biology (2024)
-
Regulatory T cells in skin regeneration and wound healing
Military Medical Research (2023)
-
ALKBH5-mediated m6A demethylation fuels cutaneous wound re-epithelialization by enhancing PELI2 mRNA stability
Inflammation and Regeneration (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.