Abstract
Diabetes is far more prevalent in smokers than non-smokers, but the underlying mechanisms of vulnerability are unknown. Here we show that the diabetes-associated gene Tcf7l2 is densely expressed in the medial habenula (mHb) region of the rodent brain, where it regulates the function of nicotinic acetylcholine receptors. Inhibition of TCF7L2 signalling in the mHb increases nicotine intake in mice and rats. Nicotine increases levels of blood glucose by TCF7L2-dependent stimulation of the mHb. Virus-tracing experiments identify a polysynaptic connection from the mHb to the pancreas, and wild-type rats with a history of nicotine consumption show increased circulating levels of glucagon and insulin, and diabetes-like dysregulation of blood glucose homeostasis. By contrast, mutant Tcf7l2 rats are resistant to these actions of nicotine. Our findings suggest that TCF7L2 regulates the stimulatory actions of nicotine on a habenula–pancreas axis that links the addictive properties of nicotine to its diabetes-promoting actions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The RNA-seq data generated in this study are available at the Gene Expression Omnibus (GEO) under the accession code GSE137118. Other data that support the findings of this study are available as Extended Data and Supplementary Information, including uncropped western blot images.
References
Stolerman, I. P. & Jarvis, M. J. The scientific case that nicotine is addictive. Psychopharmacology (Berl.) 117, 2–10, discussion 14–20 (1995).
Maskos, U. et al. Nicotine reinforcement and cognition restored by targeted expression of nicotinic receptors. Nature 436, 103–107 (2005).
Fowler, C. D., Lu, Q., Johnson, P. M., Marks, M. J. & Kenny, P. J. Habenular α5 nicotinic receptor subunit signalling controls nicotine intake. Nature 471, 597–601 (2011).
Fowler, C. D. & Kenny, P. J. Nicotine aversion: neurobiological mechanisms and relevance to tobacco dependence vulnerability. Neuropharmacology 76 Pt B, 533–544 (2014).
Tuesta, L. M. et al. GLP-1 acts on habenular avoidance circuits to control nicotine intake. Nat. Neurosci. 20, 708–716 (2017).
Liu, Z. & Habener, J. F. Glucagon-like peptide-1 activation of TCF7L2-dependent Wnt signaling enhances pancreatic beta cell proliferation. J. Biol. Chem. 283, 8723–8735 (2008).
Grant, S. F. et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat. Genet. 38, 320–323 (2006).
Sladek, R. et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445, 881–885 (2007).
Fuchsberger, C. et al. The genetic architecture of type 2 diabetes. Nature 536, 41–47 (2016).
Haggard, H. W. & Greenberg, L. A. The effects of cigarette smoking upon the blood sugar. Science 79, 165–166 (1934).
Sandberg, H., Roman, L., Zavodnick, J. & Kupers, N. The effect of smoking on serum somatotropin, immunoreactive insulin and blood glucose levels of young adult males. J. Pharmacol. Exp. Ther. 184, 787–791 (1973).
GBD 2015 Tobacco Collaborators. Smoking prevalence and attributable disease burden in 195 countries and territories, 1990-2015: a systematic analysis from the Global Burden of Disease Study 2015. Lancet 389, 1885–1906 (2017).
Willi, C., Bodenmann, P., Ghali, W. A., Faris, P. D. & Cornuz, J. Active smoking and the risk of type 2 diabetes: a systematic review and meta-analysis. J. Am. Med. Assoc. 298, 2654–2664 (2007).
Ren, J. et al. Habenula “cholinergic” neurons co-release glutamate and acetylcholine and activate postsynaptic neurons via distinct transmission modes. Neuron 69, 445–452 (2011).
Görlich, A. et al. Reexposure to nicotine during withdrawal increases the pacemaking activity of cholinergic habenular neurons. Proc. Natl Acad. Sci. USA 110, 17077–17082 (2013).
Dougherty, J. D., Schmidt, E. F., Nakajima, M. & Heintz, N. Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res. 38, 4218–4230 (2010).
Ables, J. L. et al. Retrograde inhibition by a specific subset of interpeduncular α5 nicotinic neurons regulates nicotine preference. Proc. Natl Acad. Sci. USA 114, 13012–13017 (2017).
Boj, S. F. et al. Diabetes risk gene and Wnt effector Tcf7l2/TCF4 controls hepatic response to perinatal and adult metabolic demand. Cell 151, 1595–1607 (2012).
Korinek, V. et al. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 19, 379–383 (1998).
Geurts, A. M. et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science 325, 433 (2009).
Graham, T. A., Ferkey, D. M., Mao, F., Kimelman, D. & Xu, W. Tcf4 can specifically recognize β-catenin using alternative conformations. Nat. Struct. Biol. 8, 1048–1052 (2001).
Molenaar, M. et al. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell 86, 391–399 (1996).
Ip, W., Shao, W., Chiang, Y. T. & Jin, T. The Wnt signaling pathway effector TCF7L2 is upregulated by insulin and represses hepatic gluconeogenesis. Am. J. Physiol. Endocrinol. Metab. 303, E1166–E1176 (2012).
Huang, S. M. et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620 (2009).
McGehee, D. S., Heath, M. J., Gelber, S., Devay, P. & Role, L. W. Nicotine enhancement of fast excitatory synaptic transmission in CNS by presynaptic receptors. Science 269, 1692–1696 (1995).
Zoli, M., Léna, C., Picciotto, M. R. & Changeux, J. P. Identification of four classes of brain nicotinic receptors using β2 mutant mice. J. Neurosci. 18, 4461–4472 (1998).
Murray, K. D., Choudary, P. V. & Jones, E. G. Nucleus- and cell-specific gene expression in monkey thalamus. Proc. Natl Acad. Sci. USA 104, 1989–1994 (2007).
Skoglund, G., Hussain, M. A. & Holz, G. G. Glucagon-like peptide 1 stimulates insulin gene promoter activity by protein kinase A-independent activation of the rat insulin I gene cAMP response element. Diabetes 49, 1156–1164 (2000).
Giniatullin, R., Nistri, A. & Yakel, J. L. Desensitization of nicotinic ACh receptors: shaping cholinergic signaling. Trends Neurosci. 28, 371–378 (2005).
Paradiso, K. & Brehm, P. Long-term desensitization of nicotinic acetylcholine receptors is regulated via protein kinase A-mediated phosphorylation. J. Neurosci. 18, 9227–9237 (1998).
Huganir, R. L., Delcour, A. H., Greengard, P. & Hess, G. P. Phosphorylation of the nicotinic acetylcholine receptor regulates its rate of desensitization. Nature 321, 774–776 (1986).
Li, Y. F. et al. Antidepressant- and anxiolytic-like effects of the phosphodiesterase-4 inhibitor rolipram on behavior depend on cyclic AMP response element binding protein-mediated neurogenesis in the hippocampus. Neuropsychopharmacology 34, 2404–2419 (2009).
Zhao, T. J. et al. Ghrelin secretion stimulated by β1-adrenergic receptors in cultured ghrelinoma cells and in fasted mice. Proc. Natl Acad. Sci. USA 107, 15868–15873 (2010).
Neil-Dwyer, G., Bartlett, J., McAinsh, J. & Cruickshank, J. M. β-adrenoceptor blockers and the blood–brian barrier. Br. J. Clin. Pharmacol. 11, 549–553 (1981).
O’Dell, L. E. & Nazarian, A. Enhanced vulnerability to tobacco use in persons with diabetes: a behavioral and neurobiological framework. Prog. Neuropsychopharmacol. Biol. Psychiatry 65, 288–296 (2016).
Perkins, K. A., Epstein, L. H., Sexton, J. E. & Pastor, S. Effects of smoking cessation on consumption of alcohol and sweet, high-fat foods. J. Subst. Abuse 2, 287–297 (1990).
Yamaguchi, T., Danjo, T., Pastan, I., Hikida, T. & Nakanishi, S. Distinct roles of segregated transmission of the septo-habenular pathway in anxiety and fear. Neuron 78, 537–544 (2013).
Soria-Gómez, E. et al. Habenular CB1 receptors control the expression of aversive memories. Neuron 88, 306–313 (2015).
Zhang, J. et al. Presynaptic excitation via GABAB receptors in habenula cholinergic neurons regulates fear memory expression. Cell 166, 716–728 (2016).
Chou, M. Y. et al. Social conflict resolution regulated by two dorsal habenular subregions in zebrafish. Science 352, 87–90 (2016).
Zhao, Z. et al. A central catecholaminergic circuit controls blood glucose levels during stress. Neuron 95, 138–152.e5 (2017).
Wang, Z. & Ma’ayan, A. An open RNA-Seq data analysis pipeline tutorial with an example of reprocessing data from a recent Zika virus study F100Res. 5, 1574 (2016).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Clark, N. R. et al. The characteristic direction: a geometrical approach to identify differentially expressed genes. BMC Bioinformatics 15, 79 (2014).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44 (W1), W90–W97 (2016).
Acknowledgements
This work was supported by grants from the National Institute on Drug Abuse to P.J.K. (DA020686) and I.I-T. (DA035756). We thank S. Stanley for pRV-GFP virus, J. Lindstrom for the α4β2α5 nAChR cell line, M. Conkright for the EVX1-CREB-luciferase-GFP reporter, A. Stewart for INS-1 cells and M. Hayes for the AAV1-shGlp1r-GFP virus. The Lenti-7xTcf-FFluc-SV40-mCherry reporter (7TFC) and dominant-negative Tcf7l2 construct (EdTc) were gifts from R. Nusse. Oxycodone and cocaine were supplied by the NIDA Drug Supply Program.
Author information
Authors and Affiliations
Contributions
A.D., M.P.H., M.V.M.DiB., S.P.B.C., W.M.H., P.B., R.M.O.C., M.W., C.F. and K.S.E. performed all behavioural experiments; M.I. performed electrophysiological recordings; S.P.B.C. designed and validated gRNAs; X.L. and Z.C. performed virus tracing; Q.L. and T.M.K. performed cell culture experiments; J.L.A. and I.I-T. generated and analysed the TRAP data; A.D. and Q.L. generated the RNA-seq data from wild-type and mutant rats; Z.W. and A.M. analysed RNA-seq data; H.C.O’N. performed the rubidium efflux experiments; A.M.G. generated the Tcf7l2 mutant rats; P.J.K. designed the experiments; A.D. and P.J.K. analysed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
P.J.K. is co-founder of Eolas Therapeutics Inc., which has a licensing agreement with AstraZeneca to develop small molecule treatments for drug dependence. P.J.K. has research support from Eli Lilly and Takeda USA.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peer review information Nature thanks Peter Kalivas, Tamas Horvath and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Fig. 1 Generation of Tcf7l2 mutant rats.
a, Schematic of the Rattus norvegicus Tcf7l2 gene. Exons are spliced to generate Tcf7l2 mRNA (NCBI reference sequence: NM_001191052.1). Primers for genotyping and Sanger sequencing are indicated by arrows flanking exon 5. b, Sequencing chromatograph of the Tcf7l2 mutant allele. The site of the 169-bp deletion from exon 5 and the following intron is labelled. c, Illustration of TCF7L2 wild-type protein, containing an N-terminal β-catenin binding domain (blue) and C-terminal DNA binding domain (red). Predicted open reading frames and truncated proteins generated from the Tcf7l2 mutant mRNA. Green regions on predicted truncated proteins denote ectopic amino acid sequences not found in wild-type TCF7L2 protein. d, Genotyping of wild-type and mutant Tcf7l2 rats: wild-type animal (+/+) with single band at 304 bp; heterozygous animal (+/−) with bands at 304 and 144 bp; and mutant animal (−/−) with a single band at 144 bp. Image is representative of genotyping results obtained for wild-type and mutant Tcf7l2 rats used each experiment. e, Graphical representation of mHb in coronal slice of rat brain. Image adapted from the Allen Brain Reference Atlas. f, Nissl staining showed similar mHb volumes in wild-type and mutant Tc7l2 rats. Image is representative of results obtained in three biologically independent animals from each genotype. g, Diffusion tensor imaging tractography of the fasciculus retroflexus in wild-type (n = 3) and mutant (n = 5) Tcf7l2 rats. h, Fractional anisotropy showed similar integrity (left and right sides) of the fasciculus retroflexus in wild-type (n = 3) and mutant (n = 5) Tcf7l2 rats (‘genotype’: F1, 6 = 0.000003; P = 0.99; ‘brain side’: F1, 6 = 2.562, P = 0.16; ‘genotype × brain side’: F1, 6 = 0.0007, P = 0.98). i, The frequency at different steps of positive current used to calculate the slope of the input–output curve from dorsal mHb neurons. Example traces showing typical current steps at −20, 0 and 40 pA in dorsal mHb neurons from wild-type and mutant Tcf7l2 rats. j, Input–output curve in dorsal mHb neurons from wild-type and mutant Tcf7l2 (n = 16 cells from 5 rats) rats. k, The frequency at different steps of positive current used to calculate the slope of the input-output curve from ventral mHb neurons. l, Input–output curve in ventral mHb neurons from wild-type and mutant Tcf7l2 (n = 16 cells, 5 rats) rats. m, Input resistance from mHb neurons from wild-type (13 cells, 4 rats) and mutant (16 cells, 5 rats) Tcf7l2 rats (P = 0.1036, unpaired two-tailed t-test). n, Afterhyperpolarization in mHb neurons from wild-type (13 cells, 4 rats) and mutant (16 cells, 5 rats) Tcf7l2 rats; P = 0.3043, unpaired two-tailed t-test. o, Sag current in mHb neurons wild-type (13 cells, 4 rats) and mutant (17 cells, 5 rats) Tcf7l2 rats (P = 0.1386, unpaired two-tailed t-test). p, Total distance travelled by drug-naive wild-type (n = 6) and mutant (n = 5) Tcf7l2 rats during a 60 min session. q, Total distance travelled by wild-type (n = 6) and mutant (n = 5) Tcf7l2 rats after daily injections of saline or nicotine (0.4 mg kg−1) (15 min pre-treatment time). r, Responses to the training dose of nicotine (0.03 mg kg−1 per infusion) were assessed in a group of wild-type (n = 9) and mutant (n = 11) Tcf7l2 rats on days 1 and 35 of access. Nicotine responses were similar between the wild-type and mutant Tcf7l2 rats on day 1 of access, but mutant Tcf7l2 rats escalated their intake such that their responses were higher on day 35 compared with wild-type Tcf7l2 rats, and compared with their own intake on day 1 (F1, 18 = 30.8, ****P < 0.0001, interaction effect between ‘genotype’ and ‘session’ in two-way ANOVA). Box plots show minimum–maximum range. Data are mean ± s.e.m.
Extended Data Fig. 2 CRISPR cleavage of Tcf7l2.
a, Exon diagram of mouse Tcf7l2 with the two pertinent domains highlighted and the sgRNA targeting locus. b, Bioinformatic comparison of the five different sgRNAs tested against Tcf7l2. MM, mismatches. c, Genomic cleavage percentage in mouse N2a cells of the five sgRNAs targeted against Tcf7l2. Data represent n = 3 biologically independent samples. d, T7-endonuclease-based assay illustrating intact PCR and cleaved bands of Tcf7l2 via CRISPR gene editing. Observations are from a single experiment. e, tdTomato expression in N2a cells 48 h after transduction of the AAV carrying sgRNA against Tcf7l2 (AAV-sgRNA-Tcf7l2). Data are representative of three biologically independent samples. f, Relative expression of Tcf7l2 transcripts in the N2a cells transfected with AAV-sgRNA-Tcf7l2, AAV-sgRNA-eGFP and AAV-CMV-spCas9 (Vector Biolabs) (***P < 0.001, unpaired two-tailed t-test). Data represent n = 5 biologically independent samples for each gRNA. g, Relative mRNA expression of habenular Tcf7l2 6 weeks after viral stereotaxic injections of AAV-sgRNA-Tcf7l2, AAV-sgRNA-eGFP or AAV2-hSYN1-iCre into the mHb of Rosa26LSL-spCas9-eGFP mice. Data represent n = 4 biologically independent samples for each gRNA. h, In vivo estimation of genomic cleavage of habenular Tcf7l2 6 weeks after viral stereotaxic injection of AAV-sgRNA-Tcf7l2, AAV-sgRNA-eGFP or AAV2-hSYN1-iCre in Rosa26LSL-spCas9-eGFP mice. Genomic cleavage efficiency was estimated by average re-annealed mismatches in a T7 endonuclease assay (***P < 0.001, unpaired two-tailed t-test). Data represent n = 3 biologically independent animals for each gRNA. i, Left, representative DAPI-counterstained brain slice showing Cas9-eGFP (green) and Tcf7l2 gRNA (red) targeted to the mHb of Rosa26LSL-spCas9-eGFP mice. Right, whole image of brain slice from which left panels are derived. Representative result from n = 3 mice. j, Medial habenula from Rosa26LSL-spCas9-eGFP mice injected with AAV-sgRNA-eGFP (n = 6 independent mice) or AAV-sgRNA-Tcf7l2 (n = 7 independent mice) was dissected and DNA amplicons of targeted region of Tcf7l2 sequenced. Percentage of indels detected in the targeted region of Tcf7l2 is shown (****P < 0.0001, unpaired two-tailed t-test). Coronal brain image adapted from the Allen Brain Reference Atlas. k, Donut graph showing Cas9-induced modifications to Tcf7l2 in the mHb of Rosa26LSL-spCas9-eGFP mice treated with AAV-sgRNA-Tcf7l2 (percentage of total amplicons sequenced). A total of 13 amplicons (n = 6 from AAV-sgRNA-eGFP-treated mice and n = 7 from AAV-sgRNA-Tcf7l2-treated mice) were sequenced. Data are mean ± s.e.m.
Extended Data Fig. 3 Mechanism by which TCF7L2 regulates nAChR function.
a, Effects of intra-mHb infusion of vehicle or EX-4 (12.5–100 ng) on nicotine intake in rats (n = 10) (F1.696, 15.26 = 38.3, P < 0.0001, one-way repeated measures ANOVA; **P < 0.01, ***P < 0.001, Bonferroni’s multiple comparisons test). b, Effects of intra-mHb infusion of vehicle or EX-4 (100 nM) on the latency to earn the first and second nicotine infusion of a self-administration session in rats (n = 10) (F1, 29 = 311.4, P < 0.0001, main effect of ‘infusion number’; F1, 29 = 125.4, P < 0.0001, main effect of ‘EX-4’ F1, 29 = 126.5, ****P < 0.0001, interaction effect; two-way ANOVA). c, Numbers of rats (n = 10 in total) that responded for the first and second nicotine infusion of a self-administration session after intra-mHb infusion of vehicle or EX-4 (12.5–100 ng). d, Effects of intra-mHb infusion of vehicle or rDKK1 (100 ng per side) on nicotine intake in rats (n = 11) (P = 0.45; unpaired two-tailed t-test). e, Effects of intra-mHb infusion of vehicle or XAV939 (12.5 ng per side) on nicotine intake in rats (n = 10) (P = 0.29; unpaired two-tailed t-test). f, Effects of intra-mHb infusion of vehicle or insulin (12.5 ng per side) on nicotine intake in rats (n = 13) (P = 0.29; unpaired two-tailed t-test). g–i, LiCl (g) and constitutively active-β-catenin (CA-β-catenin) (h), but not nicotine (i), increased GFP relative to mCherry expression in PC12 cells transfected with the 7xTcf-eGFP//SV40-mCherry (7TGC) TCF7L2 reporter. Data reflect results from two independent experiments. j, Levels of β-catenin phosphorylated at serine residue 675 or 552 in rat PC12 cells after treatment with forskolin, WNT3A or nicotine. Data reflect results from two independent experiments. k, LacZ expression in the mHb of BAT-GAL β-galactosidase reporter mice after nicotine injection. Data reflect results from two independent animals in each group. l, Expression levels of TCF7L2 (about 69 kDa) in the habenula were measured by western blotting in rats that responded for intravenous nicotine infusions (0.18 mg kg−1 per infusion; n = 12) or food rewards (n = 12). Each lane contains pooled tissues from n = 3 animals. Experiment was performed on a single occasion. For uncropped gel image, see Supplementary Fig. 1. m, siRNA-mediated knockdown of TCF7L2 attenuated intracellular calcium transients induced by nicotine (20 nM–320 μM) in HEK293T cells heterologously expressing α5α4β2 nAChRs (two-way repeated measures ANOVA; ‘siRNA’: F1, 4 = 63.38, P < 0.005; ‘nicotine’: F15, 60 = 1388, P < 0.0001; ‘siRNA × nicotine’: F15, 60 = 20.89, ***P < 0.0001; Bonferroni post hoc test after interaction effect in two-way ANOVA). Representative result from three experiments. n, 86Rb+ efflux from synaptosomes generated from IPn tissues derived from wild-type (n = 6) and mutant (n = 6) Tcf7l2 rats (F1,39 = 4.267, *P = 0.045; extra sum-of-squares F test). Shift in half-maximal effective concentration (EC50) value between genotypes using comparison of fits in a nonlinear fit model. o, Pharmacologically isolated nAChR currents (normalized) evoked by nicotine (0.1 Hz application) in mHb neurons from wild-type rats (n = 3 cells from 1 rat) were rapidly and completely blocked by bath application of mecamylamine (10 μM). *F1.335, 2.670 = 332.5; P < 0.001; one-way repeated measures ANOVA. p, Baseline nAChR currents in mHb neurons from wild-type (n = 13 cells, 4 rats) and mutant (n = 15 cells, 4 rats) Tcf7l2rats. P = 0.8180, unpaired two-tailed t-test. q, nAChR current decay time after nicotine stimulation (0.1 Hz) before and after nicotine (1 Hz)-induced desensitization in mHb neurons from wild-type (n = 13 cells, 4 rats) and mutant (n = 9 cells, 4 rats) Tcf7l2 rats; P = 0.7133, unpaired two-tailed t-test. r, Slope of nAChR current decay after nicotine stimulation (0.1 Hz) before and after nicotine (1 Hz)-induced desensitization in mHb neurons from wild-type (n = 13 cells, 4 rats) and mutant (n = 9 cells, 4 rats) Tcf7l2 rats; P = 0.645, unpaired two-tailed t-test. Data are mean ± s.e.m.
Extended Data Fig. 4 Genes regulated by TCF7L2 in the mHb.
a, qPCR analysis Akap9 transcript levels in the mHb of wild-type (n = 8) and mutant (n = 7) Tcf7l2 rats; and Arhgap5 transcript levels in the mHb of wild-type (n = 5) and mutant (n = 7) Tcf7l2 rats (***P < 0.001, unpaired two-tailed t-test). b, qPCR analysis of α5, α3 and β4 nAChR subunit expression in the mHb of wild-type (n = 5) and mutant (n = 8) Tcf7l2 rats. c, Top 15 most abundantly expressed genes in the mHb of wild-type Tcf7l2 rats that show differential downregulation in the mHb of mutant Tcf7l2 rats (n = 9 rats per genotype). Genes are organized in descending order according to the baseline expression levels in wild-type rats. The log2-transformed fold change of downregulated gene expression in the mHb of mutant compared with wild-type Tcf7l2 rats is shown. BaseMean, mean of normalized counts for all samples; lfcSE, s.e.m. of log2-transformed fold change; stat, Wald chi-squared test of normalized counts for gene transcript in mutant versus wild-type Tcf7l2 rats; pval, uncorrected Wald test P value; padj, P value adjusted for multiple testing using Benjamini–Hochberg to estimate the false discovery rate. d–h, Knockdown of PAFAHIB1 (d), NDFIIP1 (e), ARHGAP5 (f), HNRNPU (g) or AKAP9 (h) mRNA transcripts using a pool of validated siRNAs had no effects on nicotine-stimulated increases in [Ca2+]i in human HEK293T cells stably expressing α5α4β2 nAChRs. Data represent n = 3 biologically independent samples. Data are mean ± s.e.m.
Extended Data Fig 5 TCF7L2 regulates cAMP signalling in PC12 cells.
a, Expression of dominant-negative Tcf7l2 (dnTcf7l2) in PC12 cells reduced the activity of a cAMP-responsive luciferase reporter (EVX-1 luciferase). ***P < 0.001, unpaired two-tailed t-test. Data represent biologically independent samples from cells transfected with mCherry (n = 7) or dnTcf7l2 (n = 8). FFL, firefly luciferase; RFU, relative fluorescent units; RL, Renilla luciferase. b, dnTcf7l2 reduced baseline and EX-4-induced increases in a cAMP-responsive reporter assay in PC12 cells (F1, 10 = 19.16, **P < 0.0014, main effect of ‘dnTcf7l2’; F1, 10 = 21.31, P = 0.001, main effect of ‘EX-4’; F1, 10 = 0.027, P = 0.87, ‘dnTcf7l2 × EX-4’ interaction; two-way ANOVA). Data represent biologically independent samples from control cells (n = 3 samples), control cells treated with EX-4 (n = 3 samples), cells transfected dnTcf7l2 (n = 4 samples) and cells transfected dnTcf7l2 and treated with EX-4 (n = 4 samples). c, dnTcf7l2 reduced baseline and EX-4-evoked increases in EVX-1-luciferase in INS-1 cells, an immortalized rat pancreatic β cell line that constitutively expresses GLP-1 receptors. Data represent results from a single experiment. d, cAMP content of mHb, IPn and hippocampus were analysed in tissues from wild-type and mutant Tcf7l2 rats. Each sample contained mHb tissue from 3 rats, and data are from 4 independent samples were analysed for a total of 12 rats per genotype. e, Vector map for the pGF-CREB-mCMV-dscGFP-P2A-luciferase (CREB reporter) lentivirus. f, Brain slices containing the mHb from wild-type and mutant Tcf7l2 rats injected with CREB reporter lentivirus into mHb and injected with luciferin just before brain collection. g, EX-4 increased luciferase activity in the mHb of wild-type (n = 3) but not mutant (n = 4) Tcf7l2 rats (F1, 11 = 9.398, P = 0.0107, main effect of ‘genotype’; F6, 66 = 7.945, ***P < 0.0001, interaction effect between ‘genotype’ and ‘EX-4’; two-way repeated-measures ANOVA). h, Pre-incubation (30 min) of HEK293T cells stably expressing α5α4β2 nAChRs with nicotine (0.1–10 μM) decreased the ability of acetylcholine (0.1 mM) to stimulate increases in [Ca2+]i (F3, 12 = 188.1, P < 0.0001, main effect of ‘nicotine’ on one-way ANOVA). Data represent n = 4 independent experiment. i, 8-Br-cAMP (100–500 μM) attenuated the inhibitory effects of nicotine (0.1 μM) preincubation (30 min) on acetylcholine (0.1 mM) evoked in increases in [Ca2+]i in α5α4β2 nAChR HEK293T cells (F1, 24 = 41.20, P < 0.0001, main effect of ‘cAMP’ in two-way ANOVA). Data represent n = 5 independent experiments. Data are mean ± s.e.m.
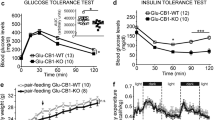
Extended Data Fig. 6 Hyperglycaemic actions of nicotine.
a, Venn diagram of differentially upregulated genes in the hippocampus, mHb and IPn of mutant Tcf7l2 rats compared with wild-type rats. b, KEGG analysis of differentially upregulated genes identified processes relevant to glucose metabolism as those most likely to be perturbed in the mHb of mutant Tcf7l2 rats (n = 9) compared with wild-type rats (n = 9). P values determined by Fisher exact test. c, Blood glucose was measured before (T0) and 30 min after (T30) rats (n = 7) were injected with saline or nicotine (1 mg kg−1) (F1, 13 = 52.3, ***P < 0.0001, interaction effect between ‘nicotine’ and ‘time’; two-way repeated-measures ANOVA). d, Oxycodone (2.5 mg kg−1) or cocaine (20 mg kg−1) injection had no effects on blood glucose in rats (n = 36). e, Chrna5-cre mice were injected into the IPn with FLEX-GFP (n = 3) or FLEX-hM3Dq-mCherry (n = 9). Image adapted from the Allen Brain Reference Atlas. f, Blood glucose was measured in both groups of mice before and 30 min after injection of CNO (1 mg kg−1); **P < 0.0051, unpaired two-tailed t-test. g, Tcf7l2 mRNA expression was reduced in the mHb of rats after shRNA-mediated knockdown of Glpr1 transcript expression. **P < 0.0051, unpaired two-tailed t-test. h, Atenolol abolished the hyperglycaemic response to experimenter-administered nicotine injection (1 mg kg−1) in rats (n = 8) (‘atenolol’, F3, 25 = 43.54, P < 0.0001; ‘time’, F2.406, 60.15 = 48.69, P < 0.0001; ‘atenolol × time’ interaction, F9, 75 = 26.88; ***P < 0.0001; two-way ANOVA). i, ICI118,551 abolished the increases in blood glucose induced by experimenter-administered nicotine injection (1 mg kg−1) in rats (n = 8) (‘nicotine’, F1, 7 = 50.83, P = 0.002; ‘ICI118,551’, F1, 7 = 13.17, P = 0.0084; ‘ICI118,551 × nicotine’ interaction, F1, 7 = 27.75, **P = 0.0012; two-way repeated-measured ANOVA). j, Atenolol abolished the increases in blood glucose induced by CNO (3 mg kg−1) in rats (n = 8) expressing FLEX-hM3Dq in the mHb–IPn circuit (‘CNO’, F1, 8 = 213.0, P < 0.0001; ‘atenolol’, F1, 8 = 27.00, P = 0.0008; ‘CNO × atenolol’ interaction, F1, 8 = 255.5, ***P < 0.0001; two-way repeated-measures ANOVA). k, Immunostaining for insulin (left), glucagon (middle) and their overlap (right) in mice treated acutely with saline (top; n = 3) or nicotine (0.5 mg kg−1; bottom; n = 3). l–o, Quantification of insulin intensity (l), insulin relative area (m), glucagon intensity (n) and glucagon relative area (o) in pancreatic islets from the saline-treated (n = 3) and nicotine-treated (n = 3) mice (**P = 0.0059, ***P < 0.001 t-test, unpaired two-tailed t-test). Image is representative of results obtained in from three biologically independent animals in each treatment group. Data are mean ± s.e.m.
Extended Data Fig. 7 Chemogenetic stimulation of the habenula.
a, Rats were injected with AAV-retro-Cre into the IPn and FLEX-GFP or FLEX-hM3Dq-mCherry into the mHb. mCherry-positive cells were detected in the mHb, confirming that virus targeting was effective. b, CNO (10 μM) had no effects on the relative spike frequency of mCherry-negative cells (n = 4 cells, 2 rats). c, CNO (10 μM) increased the relative spike frequency of mCherry-positive cells (n = 4 cells, 3 rats) (*P = 0.0124, unpaired two-tailed t-test). d, Nicotine (1 μM) increased the relative spike frequency of mHb neurons by a magnitude similar to that seen in mCherry-positive neurons after CNO treatment (n = 6 cells, 3 rats) (*P = 0.042, unpaired two-tailed t-test). Data are mean ± s.e.m.
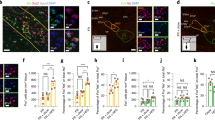
Extended Data Fig. 8 pRV mapping of polysynaptic projections from brain to pancreas and liver.
a, c, Images of a pRV-GFP-labelled cells (indicated by white arrows) and fibres in the mHb. b, d, Representative images of pRV-GFP-labelled IPn neurons (indicated by white arrows). e, Images of GFP-labelled cells in hypothalamus, cortex, substantia nigra and nucleus of the solitary tract (NTS) after pancreas injection of pRV-GFP. r, Images of a GFP-labelled cells in hypothalamus, ventral tegmental area (VTA) and NTS after liver injection of pRV-GFP. Note the absence of GFP-positive cells in the medial habenula. Images are representative of results obtained from three separate experiments. Data are mean ± s.e.m.
Extended Data Fig. 9 Consequences of hyperglycaemic actions of nicotine.
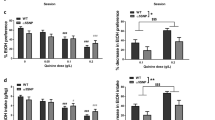
a, Effects of glucose (1 mg kg−1, intravenous) on nicotine (0.03 mg kg-1 per infusion) intake in rats (n = 15). b, Effects of glucose (1 mg kg−1, intravenous) on nicotine (0.12 mg kg-1 per infusion) intake in rats (n = 16). c, Effects of glucose (2 mg kg−1, orally) on nicotine (0.12 mg kg-1 per infusion) intake in rats (n = 16). d, Effects of glucagon (0.2 mg kg−1, intravenous) on nicotine (0.12 mg kg−1 per infusion) intake in rats (n = 5). e, Atenolol (10 mg kg−1) delivered before the self-administration on three consecutive days did not alter nicotine (0.12 mg kg−1 per infusion) intake in rats (n = 8). f, Scatter plots of average TRAP immunoprecipitation samples from sucrose-drinking (y axis; n = 28) versus immunoprecipitation samples from sucrose-naive (x axis; n = 8) ChATDW167 mice representing increased (>0.5 log2-transformed fold change, magenta) or decreased (< −0.5 log2-transformed fold change, blue) levels of transcripts undergoing translation (tissues from n = 4 mice were pooled for each sample; 7 samples from sucrose-drinking and 2 samples from sucrose-naive mice were used). Differentially expressed genes were identified by performing a negative binomial test using DESeq2, with default settings. Significant P values were corrected to control the false discovery rate of multiple testing according to the Benjamini–Hochberg procedure at 0.05 threshold and minimum threshold of 0.6 log2-transformed fold change. g, Expression levels (z-score-transformed normalized counts) of the top 50 genes affected by sucrose consumption in mHb cholinergic neurons. h, KEGG analysis of differentially upregulated genes in the mHb of ChATDW167 mice described in f identified pathways that are likely to be affected in the mHb by sucrose consumption. P values determined by Fisher exact test. i, KEGG analysis of differentially downregulated genes in the mHb of ChATDW167 mice described in f identified pathways that are likely to be affected in the mHb by sucrose consumption. P values determined by Fisher exact test. j, The frequency of action potentials in mHb neurons was unaltered by increasing the glucose concentrations in the extracellular solution from 12.5 to 30 mM (n = 6 cells from 3 rats). k, Maintaining glucose concentration in artificial cerebrospinal fluid (aCSF) in the extracellular solution at 12.5 mM did not alter the magnitude of nicotine-evoked nAChR currents in mHb neurons (n = 6 cells from 3 rats). l, Increasing the glucose concentration in the extracellular solution from 12.5 to 30 mM decreased the magnitude of nicotine (1 μM) evoked nAChR currents in mHb neurons (n = 6 cells from 3 rats) (*P < 0.0121, unpaired two-tailed t-test). m, Blood glucose levels measured in rats 24 h after their final nicotine (0.12 mg kg−1 per infusion; n = 7) or saline (n = 8) self-administration session (*P < 0.0223, unpaired two-tailed t-test). n, Blood glucose levels measured in rats 6 weeks after their final nicotine (0.12 mg kg−1 per infusion; n = 7) or saline (n = 8) self-administration session (*P < 0.0371, unpaired two-tailed t-test). o, Body weights in post-saline (n = 8) and post-nicotine rats (n = 6) measured 6 weeks after their final self-administration session. p, Fasting blood glucose levels in wild-type (n = 14 in total) and mutant (n = 14 in total) Tcf7l2 rats measured before chronic saline or nicotine injections commenced. q, Circulating levels of glucagon in nicotine-naive wild-type (n = 7) and mutant (n = 7) Tcf7l2 rats. r, Circulating levels of insulin in nicotine-naive wild-type (n = 7) and mutant (n = 7) Tcf7l2 rats. s, Circulating glucagon levels in wild-type (n = 9 in total) and mutant (n = 10 in total) Tcf7l2 rats measured before chronic saline or nicotine injections ended (F1, 15 = 4.606, *P < 0.0486, interaction effect of ‘genotype × nicotine’ in two-way ANOVA). Data are mean ± s.e.m.
Extended Data Fig. 10 Proposed mechanism by which TCF7L2 regulates the motivational properties of nicotine and its disruptive effects on blood glucose homeostasis.
A representation of a mHb neuron projecting monosynaptically to the IPn (both in blue), via the fasciculus retroflexus, and to the pancreas via a polysynaptic pathway (broken line) is shown. The mHb neurons expresses nAChRs that are activated by nicotine and that undergo nicotine-induced desensitization. In wild-type rats, nAChRs rapidly recover from desensitization by a process involving cAMP signalling. In mutant Tcf7l2 rats, cAMP signalling is compromised, which results in persistently desensitized nAChRs and diminished sensitivity of mHb neurons to nicotine. When mHb neurons are activated by nicotine, IPn neurons are stimulated by mHb-derived acetylcholine and glutamate. This triggers nicotine avoidance and a hyperglycaemic response, both of which are attenuated in mutant Tcf7l2 rats. After chronic exposure to the hyperglycaemic actions of nicotine, circulating levels of the pancreas-derived hormones glucagon and insulin are increased, resulting in a diabetes-like disruption of glucose homeostasis. This diabetes-promoting action of nicotine is also attenuated in mutant Tcf7l2 rats.
Supplementary information
Supplementary Figures
This file contains the full-length uncropped Western blot images
Rights and permissions
About this article
Cite this article
Duncan, A., Heyer, M.P., Ishikawa, M. et al. Habenular TCF7L2 links nicotine addiction to diabetes. Nature 574, 372–377 (2019). https://doi.org/10.1038/s41586-019-1653-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1653-x
This article is cited by
-
Generative artificial intelligence-enabled dynamic detection of rat nicotine-related circuits
Neural Computing and Applications (2024)
-
Insulin Resistance/Diabetes and Schizophrenia: Potential Shared Genetic Factors and Implications for Better Management of Patients with Schizophrenia
CNS Drugs (2024)
-
The controversial effect of smoking and nicotine in SARS-CoV-2 infection
Allergy, Asthma & Clinical Immunology (2023)
-
Nicotine rebalances NAD+ homeostasis and improves aging-related symptoms in male mice by enhancing NAMPT activity
Nature Communications (2023)
-
Advanced neurobiological tools to interrogate metabolism
Nature Reviews Endocrinology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.