Abstract
Salinity is detrimental to plant growth, crop production and food security worldwide. Excess salt triggers increases in cytosolic Ca2+ concentration, which activate Ca2+-binding proteins and upregulate the Na+/H+ antiporter in order to remove Na+. Salt-induced increases in Ca2+ have long been thought to be involved in the detection of salt stress, but the molecular components of the sensing machinery remain unknown. Here, using Ca2+-imaging-based forward genetic screens, we isolated the Arabidopsis thaliana mutant monocation-induced [Ca2+]i increases 1 (moca1), and identified MOCA1 as a glucuronosyltransferase for glycosyl inositol phosphorylceramide (GIPC) sphingolipids in the plasma membrane. MOCA1 is required for salt-induced depolarization of the cell-surface potential, Ca2+ spikes and waves, Na+/H+ antiporter activation, and regulation of growth. Na+ binds to GIPCs to gate Ca2+ influx channels. This salt-sensing mechanism might imply that plasma-membrane lipids are involved in adaption to various environmental salt levels, and could be used to improve salt resistance in crops.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
References
Munns, R. & Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681 (2008).
Ismail, A. M. & Horie, T. Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu. Rev. Plant Biol. 68, 405–434 (2017).
Yang, Y. & Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217, 523–539 (2018).
Deinlein, U. et al. Plant salt-tolerance mechanisms. Trends Plant Sci. 19, 371–379 (2014).
Zhu, J. K. Abiotic stress signaling and responses in plants. Cell 167, 313–324 (2016).
Julkowska, M. M. & Testerink, C. Tuning plant signaling and growth to survive salt. Trends Plant Sci. 20, 586–594 (2015).
Kudla, J. et al. Advances and current challenges in calcium signaling. New Phytol. 218, 414–431 (2018).
Edel, K. H., Marchadier, E., Brownlee, C., Kudla, J. & Hetherington, A. M. The evolution of calcium-based signalling in plants. Curr. Biol. 27, R667–R679 (2017).
Knight, M. R., Campbell, A. K., Smith, S. M. & Trewavas, A. J. Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352, 524–526 (1991).
Knight, H., Trewavas, A. J. & Knight, M. R. Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J. 12, 1067–1078 (1997).
Hedrich, R. Ion channels in plants. Physiol. Rev. 92, 1777–1811 (2012).
Roper, S. D. & Chaudhari, N. Taste buds: cells, signals and synapses. Nat. Rev. Neurosci. 18, 485–497 (2017).
Chandrashekar, J. et al. The cells and peripheral representation of sodium taste in mice. Nature 464, 297–301 (2010).
Zhang, Y. V., Ni, J. & Montell, C. The molecular basis for attractive salt-taste coding in Drosophila. Science 340, 1334–1338 (2013).
Oka, Y., Butnaru, M., Von Buchholtz, L., Ryba, N. J. P. & Zuker, C. S. High salt recruits aversive taste pathways. Nature 494, 472–475 (2013).
Chatzigeorgiou, M., Bang, S., Hwang, S. W. & Schafer, W. R. tmc-1 encodes a sodium-sensitive channel required for salt chemosensation in C. elegans. Nature 494, 95–99 (2013).
Yuan, F. et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 514, 367–371 (2014).
Choi, W. G., Hilleary, R., Swanson, S. J., Kim, S. H. & Gilroy, S. Rapid, long-distance electrical and calcium signaling in plants. Annu. Rev. Plant Biol. 67, 287–307 (2016).
Martí, M. C., Stancombe, M. A. & Webb, A. A. R. Cell- and stimulus type-specific intracellular free Ca2+ signals in Arabidopsis. Plant Physiol. 163, 625–634 (2013).
Hedrich, R., Salvador-Recatalà, V. & Dreyer, I. Electrical wiring and long-distance plant communication. Trends Plant Sci. 21, 376–387 (2016).
Choi, W. G., Toyota, M., Kim, S. H., Hilleary, R. & Gilroy, S. Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl Acad. Sci. USA 111, 6497–6502 (2014).
Evans, M. J., Choi, W. G., Gilroy, S. & Morris, R. J. A ROS-assisted calcium wave dependent on the AtRBOHD NADPH oxidase and TPC1 cation channel propagates the systemic response to salt stress. Plant Physiol. 171, 1771–1784 (2016).
Rennie, E. A. et al. Identification of a sphingolipid α-glucuronosyltransferase that is essential for pollen function in Arabidopsis. Plant Cell 26, 3314–3325 (2014).
Rennie, E. A. et al. Three members of the Arabidopsis glycosyltransferase family 8 are xylan glucuronosyltransferases. Plant Physiol. 159, 1408–1417 (2012).
Nikolovski, N., Shliaha, P. V., Gatto, L., Dupree, P. & Lilley, K. S. Label-free protein quantification for plant Golgi protein localization and abundance. Plant Physiol. 166, 1033–1043 (2014).
Tartaglio, V. et al. Glycosylation of inositol phosphorylceramide sphingolipids is required for normal growth and reproduction in Arabidopsis. Plant J. 89, 278–290 (2017).
Gronnier, J., Germain, V., Gouguet, P., Cacas, J.-L. & Mongrand, S. GIPC: glycosyl inositol phospho ceramides, the major sphingolipids on earth. Plant Signal. Behav. 11, e1152438 (2016).
Markham, J. E., Lynch, D. V., Napier, J. A., Dunn, T. M. & Cahoon, E. B. Plant sphingolipids: function follows form. Curr. Opin. Plant Biol. 16, 350–357 (2013).
Hannun, Y. A. & Obeid, L. M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 19, 175–191 (2018).
Cacas, J. L. et al. Revisiting plant plasma membrane lipids in tobacco: a focus on sphingolipids. Plant Physiol. 170, 367–384 (2016).
Ledeen, R. W. & Wu, G. The multi-tasked life of GM1 ganglioside, a true factotum of nature. Trends Biochem. Sci. 40, 407–418 (2015).
Schnaar, R. L., Gerardy-Schahn, R. & Hildebrandt, H. Sialic acids in the brain: gangliosides and polysialic acid in nervous system development, stability, disease, and regeneration. Physiol. Rev. 94, 461–518 (2014).
Green, W. N. & Andersen, O. S. Surface charges and ion channel function. Annu. Rev. Physiol. 53, 341–359 (1991).
Liu, K. H. et al. Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 545, 311–316 (2017).
Ho, C. H., Lin, S. H., Hu, H. C. & Tsay, Y. F. CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194 (2009).
Xu, J. et al. A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125, 1347–1360 (2006).
McLaughlin, S. & Murray, D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature 438, 605–611 (2005).
Balla, T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93, 1019–1137 (2013).
Hirschi, M. et al. Cryo-electron microscopy structure of the lysosomal calcium-permeable channel TRPML3. Nature 550, 411–414 (2017).
Gao, Y., Cao, E., Julius, D. & Cheng, Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 534, 347–351 (2016).
Zipfel, C. & Oldroyd, G. E. D. Plant signalling in symbiosis and immunity. Nature 543, 328–336 (2017).
Hamilton, E. S., Schlegel, A. M. & Haswell, E. S. United in diversity: mechanosensitive ion channels in plants. Annu. Rev. Plant Biol. 66, 113–137 (2015).
Choi, J. et al. Identification of a plant receptor for extracellular ATP. Science 343, 290–294 (2014).
Ranf, S. et al. A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat. Immunol. 16, 426–433 (2015).
Charpentier, M. et al. Nuclear-localized cyclic nucleotide-gated channels mediate symbiotic calcium oscillations. Science 352, 1102–1105 (2016).
Dindas, J. et al. AUX1-mediated root hair auxin influx governs SCFTIR1/AFB-type Ca2+ signaling. Nat. Commun. 9, 1174 (2018).
Mousavi, S. A. R., Chauvin, A., Pascaud, F., Kellenberger, S. & Farmer, E. E. GLUTAMATE RECEPTOR-LIKE genes mediate leaf-to-leaf wound signalling. Nature 500, 422–426 (2013).
Ortiz-Ramírez, C. et al. GLUTAMATE RECEPTOR-LIKE channels are essential for chemotaxis and reproduction in mosses. Nature 549, 91–95 (2017).
Murthy, S. E., Dubin, A. E. & Patapoutian, A. Piezos thrive under pressure: mechanically activated ion channels in health and disease. Nat. Rev. Mol. Cell Biol. 18, 771–783 (2017).
Lenarčič, T. et al. Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins. Science 358, 1431–1434 (2017).
Monshausen, G. B., Messerli, M. A. & Gilroy, S. Imaging of the Yellow Cameleon 3.6 indicator reveals that elevations in cytosolic Ca2+ follow oscillating increases in growth in root hairs of Arabidopsis. Plant Physiol. 147, 1690–1698 (2008).
Feldmann, K. A. T-DNA insertion mutagenesis in Arabidopsis - mutational spectrum. Plant J. 1, 71–82 (1991).
Weigel, D. et al. Activation tagging in Arabidopsis. Plant Physiol. 122, 1003–1013 (2000).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Schubert, D. et al. Silencing in Arabidopsis T-DNA transformants: the predominant role of a gene-specific RNA sensing mechanism versus position effects. Plant Cell 16, 2561–2572 (2004).
Li, J. et al. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222 (2002).
Pei, Z.-M. et al. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406, 731–734 (2000).
Tang, R. H. et al. Coupling diurnal cytosolic Ca2+ oscillations to the CAS-IP3 pathway in Arabidopsis. Science 315, 1423–1426 (2007).
Lahner, B. et al. Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat. Biotechnol. 21, 1215–1221 (2003).
Baxter, I. et al. A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLoS Genet. 6, e1001193 (2010).
Song, W. Y. et al. Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proc. Natl Acad. Sci. USA 107, 21187–21192 (2010).
Qiu, Q. S., Guo, Y., Dietrich, M. A., Schumaker, K. S. & Zhu, J. K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl Acad. Sci. USA 99, 8436–8441 (2002).
Johansson, F., Olbe, M., Sommarin, M. & Larsson, C. Brij 58, a polyoxyethylene acyl ether, creates membrane vesicles of uniform sidedness. A new tool to obtain inside-out (cytoplasmic side-out) plasma membrane vesicles. Plant J. 7, 165–173 (1995).
Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976).
Blumwald, E. & Poole, R. J. Na/H antiport in isolated tonoplast vesicles from storage tissue of Beta vulgaris. Plant Physiol. 78, 163–167 (1985).
Schwacke, R. et al. ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131, 16–26 (2003).
Karimi, M., Inzé, D. & Depicker, A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 (2002).
Nelson, B. K., Cai, X. & Nebenführ, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136 (2007).
Hooper, C. M., Castleden, I. R., Tanz, S. K., Aryamanesh, N. & Millar, A. H. SUBA4: the interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res. 45, D1064–D1074 (2017).
Heard, W., Sklenář, J., Tomé, D. F. A., Robatzek, S. & Jones, A. M. E. Identification of regulatory and cargo proteins of endosomal and secretory pathways in Arabidopsis thaliana by proteomic dissection. Mol. Cell. Proteomics 14, 1796–1813 (2015).
Nikolovski, N. et al. Putative glycosyltransferases and other plant Golgi apparatus proteins are revealed by LOPIT proteomics. Plant Physiol. 160, 1037–1051 (2012).
Jefferson, R. A., Kavanagh, T. A. & Bevan, M. W. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907 (1987).
Buré, C. et al. Fast screening of highly glycosylated plant sphingolipids by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 25, 3131–3145 (2011).
Wang, P. et al. Cell membrane surface potential (ψ 0) plays a dominant role in the phytotoxicity of copper and arsenate. Plant Physiol. 148, 2134–2143 (2008).
Delgado, A. V., González-Caballero, F., Hunter, R. J., Koopal, L. K. & Lyklema, J. Measurement and interpretation of electrokinetic phenomena. J. Colloid Interface Sci. 309, 194–224 (2007).
Kinraide, T. B. & Wang, P. The surface charge density of plant cell membranes (σ): an attempt to resolve conflicting values for intrinsic σ. J. Exp. Bot. 61, 2507–2518 (2010).
Yoo, S. D., Cho, Y. H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protocols 2, 1565–1572 (2007).
Endo, M., Shimizu, H. & Araki, T. Rapid and simple isolation of vascular, epidermal and mesophyll cells from plant leaf tissue. Nat. Protocols 11, 1388–1395 (2016).
Welsch, M. E. et al. Multivalent small-molecule pan-RAS inhibitors. Cell 168, 878–889 (2017).
Menting, J. G. et al. How insulin engages its primary binding site on the insulin receptor. Nature 493, 241–245 (2013).
Coscia, F. et al. Multi-level proteomics identifies CT45 as a chemosensitivity mediator and immunotherapy target in ovarian cancer. Cell 175, 159–170 (2018).
Chen, Y. N. P. et al. Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature 535, 148–152 (2016).
Johnson, R. A., Manley, O. M., Spuches, A. M. & Grossoehme, N. E. Dissecting ITC data of metal ions binding to ligands and proteins. Biochim. Biophys. Acta. 1860, 892–901 (2016).
Machaidze, G., Ziegler, A. & Seelig, J. Specific binding of Ro 09-0198 (cinnamycin) to phosphatidylethanolamine: a thermodynamic analysis. Biochemistry 41, 1965–1971 (2002).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650 (2009).
Yin, Y., Chen, H., Hahn, M. G., Mohnen, D. & Xu, Y. Evolution and function of the plant cell wall synthesis-related glycosyltransferase family 8. Plant Physiol. 153, 1729–1746 (2010).
Acknowledgements
We thank J. Li for the pBIB-BASTA vector; M. R. Knight for aequorin-expressing seeds; S. Gilroy for YC3.6-expressing seeds; M. Chen, X. Dong and T.-p. Sun for advice on physical mapping; and T.-p. Sun, J. Pei, D. R. McClay and P. N. Benfey for discussion and reading of the manuscript. Z.J., M.T., F.W., W.Z. and H. Zhang were supported in part by the Shenzhen Peacock Innovative Research Team Program; Z.J., X.Z., L.L., X.W., F.W. and F.Y. by the Pandeng Project of Hangzhou Normal University and Chinese NSF (31301170, U1130304); and F.W., Y.N. and Y.X. by fellowships from the China Scholarship Council. This work was supported by grants from NSF (IOS-1457257, IOS-0848263) and USDA (CSREES-2006-35100-17304) to Z.-M.P.; National Key Project for Synthetic Biology (2018YFA0902500) to Z.H.; and DOE (DE-SC0014077) to T.V.-D. and Z.-M.P.
Author information
Authors and Affiliations
Contributions
Z.J., F.Y., Y. Xiang and Y.N. conducted genetic screens. Z.J., X.Z., F.Y., L.L., F.L., Y. Xue, F.W., X.W. and H. Zhao performed map-based cloning. C.L., R.Y., F.Y. and B.B. generated T-DNA-mutagenized Arabidopsis populations. Z.J., L.L., F.W., M.T., C.H., H. Zhao, C.P. and W.Z. carried out physiological analyses. Z.J., M.T., W.Z., and Z.H. analysed GIPC content and ion content. M.T., Z.J., W.Z., and Z.H. analysed ion binding to GIPCs. Z.J., C.P. and F.W. conducted zeta potential measurement. Z.J., L.L., H.-N.W., B.M.C., D.M.J., G.B.S., H. Zhang and T.V.-D. participated in Ca2+-imaging analyses. Z.-M.P. designed and supervised the overall research with input from Z.H., Z.J. and J.N.S. Z.-M.P., Z.J. and J.N.S. wrote the paper. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 The optimized conditions for genetic screens that distinguish increases in [Ca2+]i induced by salt from those induced by osmotic stress.
a, Aequorin bioluminescence imaging of [Ca2+]i induced by NaCl at concentrations of 0, 100, or 200 mM in aequorin-expressing Arabidopsis seedlings. Relative [Ca2+]i in leaves is shown using a pseudo-colour scale. Similar results were seen in more than 20 independent experiments. b, Aequorin imaging of [Ca2+]i induced by sorbitol at concentrations of 0, 200, or 400 mM (at equivalent osmolalities to 0, 100, and 200 mM NaCl in a, respectively). Seedlings grown for 7 days were treated with NaCl or sorbitol solutions at indicated concentrations, and the aequorin bioluminescence images were acquired as described for experiments in Fig. 1a. Similar results were seen in more than 20 independent experiments.
Extended Data Fig. 2 The moca1 mutant has wild-type-like growth phenotypes and shows defects in increases in [Ca2+]i induced by salt but not by osmotic stress.
a, Representative photographs of wild-type and moca1 plants grown in soil taken at 12 days (top; young seedling stage), 22 days (middle; bolting stage), and 32 days (bottom; flowering and seed setting stage). Similar results from several batches of seeds were seen in more than 50 experiments. b, c, Leaf area (b) and shoot weight (c) from 12-day-old seedlings as in a (b, P = 0.729; c, P = 0.912). d, e, Leaf area (d) and shoot weight (e) from 22-day-old plants as in a (c, P = 0.141; d, P = 0.179). f, g, Plant height (f) and shoot weight (g) from 32-day-old plants as in a (f, P = 0.054; g, P = 0.168). b–f, Mean ± s.d.; n = 18 seedlings. h, i, Similar total amount of remaining aequorin bioluminescence in wild-type and moca1 seedlings. The same seedlings used in Fig. 1b were treated with a solution containing 0.9 M CaCl2 and 10% (v/v) ethanol to measure the total amount of remaining aequorin, and no difference between wild-type and moca1 seedlings was observed (h). Similar results were seen in more than 30 separate experiments. Quantification of total amount of aequorin in wild-type and moca1 plants from experiments as in h (i; mean ± s.d.; n = 10 pools (20 seedlings per pool); P = 0.893). j, k, The moca1 mutant shows significantly lower elevation in [Ca2+]i induced by 200 mM NaCl than the wild type. Wild-type and moca1 seedlings grown side-by-side were treated with 200 mM NaCl solution (∼400 mOsm) and changes in [Ca2+]i were recorded (j). Data are quantified from experiments in j (k; mean ± s.d.; n = 16 seedlings; P < 0.001). Similar results were seen in more than 20 independent experiments. l, m, Similar increases in [Ca2+]i induced by 400 mM sorbitol (with about equal osmolality to 200 mM NaCl) in wild-type and moca1 seedlings. Wild-type and moca1 seedlings grown side-by-side were treated with 400 mM (∼400 mOsm) sorbitol solution and changes in [Ca2+]i in leaves were recorded (l). Data are quantified from experiments in l (m; mean ± s.d.; n = 16 seedlings; P = 0.141). Similar results were seen in more than 20 independent experiments.
Extended Data Fig. 3 Differences in Ca2+ signalling between salt stress-sensing moca1 and osmotic stress-sensing osca1 mutants and impaired activation of SOS1 Na+/H+ antiporter by salt in moca1.
a−c, Representative aequorin bioluminescence images of wild-type, moca1 and osca1 seedlings grown side-by-side and treated with water (a), 200 mM NaCl (b), or 400 mM sorbitol (c). Similar results were seen in more than 20 independent experiments. d, e, Averaged increases in [Ca2+]i plotted as a function of applied NaCl concentrations (d) and sorbitol concentrations (e) in wild-type, moca1 and osca1 leaves. Similar results were seen in more than ten separate experiments. Data are from three experiments (mean ± s.d.; n = 48 seedlings for each data point). f, Representative fluorescence traces of the pH-sensitive probe quinacrine show plasma membrane Na+/H+ exchange activity from wild-type and moca1 plants without (0 mM NaCl) or with salt treatment (100 mM NaCl) for 24 h. Plasma membrane vesicles were prepared, and ΔpH was established by activation of the plasma membrane H+-ATPase (MgSO4) and measured as a decrease (quench) in the fluorescence of quinacrine (see Methods). Na+/H+ exchanger activity was measured as an increase in fluorescence (NaCl; highlighted). Quantification of the results is shown in Fig. 2f. Similar results were seen in three independent experiments.
Extended Data Fig. 4 The moca1 mutant does not affect responses to osmotic, KCl or LiCl stresses or the abscisic acid signalling pathway.
a, b, Wild-type and moca1 seedlings were grown in low Ca2+ and ½ MS medium containing 0 mM or 200 mM sorbitol for 12 days (a), and root lengths were analysed (b). Data are from three independent experiments (mean ± s.d.; n = 13 pools (8 seedlings per pool); two-way ANOVA, P = 0.064). c, d, Wild-type and moca1 seedlings were grown in low Ca2+ and ½ MS medium in the presence or absence of abscisic acid (ABA) for 12 days (c), and root length was quantified (d). Data are from three independent experiments (mean ± s.d.; n = 13 pools (8–12 seedlings per pool); two-way ANOVA, P = 0.249). e–g, Wild-type and moca1 seedlings were grown in low Ca2+ and ½ MS medium containing 0–80 mM KCl for 12 days (e), and root lengths (f) and fresh weight (g) from experiments in e were quantified. Data are from three independent experiments (mean ± s.d.; n = 12 pools (8–12 seedlings per pool); f, P = 0.773 (–KCl), P = 0.707 (+KCl); g, P = 0.126 (–KCl), P = 0.281 (+KCl)). h, Wild-type and moca1 seedlings were grown in low Ca2+ and ½ MS medium containing 0 mM or 20 mM LiCl for 12 days. Severe inhibition of growth was observed in both wild-type and moca1 seedlings at 20 mM LiCl. At low concentrations of LiCl, no differences were observed. Similar results were seen in more than 10 independent experiments. In contrast to NaCl stress (Fig. 2c–e), KCl and LiCl stresses inhibited the growth of wild-type and moca1 seedlings at almost the same level (Extended Data Fig. 4e–h). Although the initial increases in [Ca2+]i and activation of the SOS pathway are almost identical for short-term Na+, K+, and Li+ stresses (Figs. 1e, 3d–f), the higher selectivity of SOS1 for Na+ over K+ and Li+ might lead to higher Na+ buildup but similar K+ and Li+ buildup in moca1 seedlings compared to wild-type seedlings. Note also that, in natural environments, plants are exposed to NaCl stress often but KCl and LiCl stress rarely, implying that the hypersensitivity of moca1 seedlings to NaCl stress is physiologically relevant.
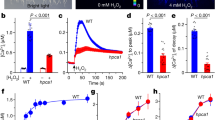
Extended Data Fig. 5 Selectivity of increases in [Ca2+]i in response to various abiotic stimuli in the moca1 mutant.
a, b, Aequorin bioluminescence image of seedlings treated with 4 mM H2O2 (a) and quantification of [Ca2+]i in leaves from experiments in a (b; P = 0.982). Aequorin images were acquired similarly to experiments in Fig. 3a. c, d, Aequorin bioluminescence image of seedlings treated with 12 °C cold water (c) and quantification of [Ca2+]i in leaves from experiments in c (d; P = 0.286). Aequorin images were acquired similarly to experiments in Fig. 3b. e, f, Aequorin bioluminescence image of seedlings treated with 135 mM CaCl2 (∼400 mOsm) (e) and quantification of [Ca2+]i in leaves from experiments in e (f; P = 0.057). Aequorin images were acquired similarly to experiments in Fig. 3c. g, h, Aequorin bioluminescence image of seedlings treated with 200 mM KCl (∼400 mOsm) (g) and quantification of [Ca2+]i in leaves from experiments in g (h; P < 0.001). Aequorin images were acquired similarly to experiments in Fig. 3d. i, j, Aequorin bioluminescence image of seedlings treated with 200 mM LiCl (∼400 mOsm) (i) and quantification of [Ca2+]i in leaves from experiments in i (j; P < 0.001). Aequorin images were acquired similarly to experiments in Fig. 3e. k, l, Aequorin bioluminescence image of seedlings treated with 200 mM NaNO3 (∼400 mOsm) (k) and quantification of [Ca2+]i in leaves from experiments in k (l; P < 0.001). Aequorin images were acquired similarly to experiments in Fig. 3f. All data show mean ± s.d.; n = 16 seedlings.
Extended Data Fig. 6 Effect of Ca2+ channel blockers on salt-induced wave-like propagation of [Ca2+]i in wild-type and moca1 roots.
a, Wave-like propagation of [Ca2+]i increases through the root. Increases in [Ca2+]i were monitored in ROIs 1,000 μm from the site of local application of 200 mM NaCl to root tip. Top, wild-type; middle, wild-type with 10 min pretreatment with 50 μM LaCl3; bottom, moca1 roots. YC3.6 Ca2+ imaging was carried out similarly to experiments in Fig. 3g–i. Similar results were seen in more than 20 independent experiments. b, Quantitative analyses of time course of [Ca2+]i changes in ROIs in response to local treatment with 200 mM NaCl in similar experiments as in a (mean ± s.d.; n = 10 ROIs). c, Peak ratio changes from experiments similar to a, b (mean ± s.d.; n = 10 ROIs).
Extended Data Fig. 7 Genetic analysis and map-based cloning of MOCA1.
a, All F1 seedlings derived from moca1 × wild-type (ColAQ, Col-0 expressing aequorin) crosses showed wild-type salt-induced increases in [Ca2+]i. F2 seedlings showed a 3:1 wild-type:moca1 segregation, suggesting that the moca1 phenotype resulted from a recessive mutation in a single nuclear gene. The F2 seedlings, which were derived from moca1 × Wassilewskija (Ws) crosses and also identified as aequorin homozygous, showed a 3:1 wild-type:moca1 segregation. The same number of F2 seeds for each cross were placed in Petri dishes and the phenotypes of salt-induced increases in [Ca2+]i were scored for individual seedlings (mean ± s.e.m.; n = 4 for moca1 × ColAQ and moca1 × Ws crosses). b, Physical mapping of MOCA1. MOCA1 was positioned between NGA249 and 5-AB006708-2862 markers in the short arm of chromosome 5 in a segregating F2 population derived from the moca1 × Ws cross. MOCA1 was fine-mapped to a region between MO59 and UPSC-5-6009 by analysing 720 recombinant chromosomes (360 lines) in the F2 population with molecular markers described in c. We sequenced all open reading frames (ORFs) in this region between these two markers and identified one deletion in an ORF, which corresponded to the gene At5g18480. c, Molecular markers developed for fine mapping. d, MOCA1 encodes a protein with six transmembrane α-helices (blue). Four amino acid residues from 493 to 496 (LMVG; red) are deleted in moca1. e, Transmembrane α-helical spanners predicted by various models using Aramemnon (http://aramemnon.botanik.uni-koeln.de).
Extended Data Fig. 8 The moca1 mutant is hypersensitive to salt stress and MOCA1 encodes a glucuronosyltransferase.
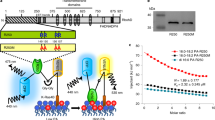
a, b, Wild-type, moca1 and complementation transgenic seedlings (MOCA1 moca1, endogenous MOCA1 promoter-driven complementation lines; MOCA1ox moca1, 35S-driven MOCA1 overexpression complementation lines) have similar total amounts of remaining aequorin. The same seedlings used in Fig. 4b were treated with a solution containing 0.9 M CaCl2 and 10% (v/v) ethanol to measure the total amount of remaining aequorin (a). Similar results were seen in ten separate experiments. The total amount of remaining aequorin in wild-type, moca1 and transgenic plants was quantified from experiments as in a (b; mean ± s.d., n = 40 pools (30 seedlings per pool)). c–e, Complementation of salt hypersensitivity of growth in the MOCA1 moca1 and MOCA1ox moca1 transgenic lines. Plants grown in medium in the absence (left) or presence of 60 mM NaCl (right) (c); and root length (d) and survival rate (e) from experiments in c (mean ± s.d.; n = 20 pools (16 seedlings per pool)). f, MOCA1 gene expressed in stamens and flowerers. GUS expression was examined using transgenic plants carrying the pMOCA1::GUS construct as in Fig. 4d. Data are representative of more than ten independent experiments. g, Golgi membrane localization of both MOCA1 and mutant MOCA1 (mMOCA1) proteins. GFP fluorescence was analysed in seedlings expressing the pMOCA1::MOCA1–GFP construct (top) or the pMOCA1::mMOCA1–GFP construct (bottom). Data are representative of more than ten independent experiments. h, Diagram of plant GIPC structure adapted from previous studies23,27,30. Hydroxyl group and net negative charge as well as the outline of MOCA1 (IPUT1)-catalysed GIPC synthesis are illustrated. Hex, hexose; GlcA, glucuronic acid; Ins, inositol; VLCFA, very long chain fatty acid; LCB, long chain base.
Extended Data Fig. 9 ITC analysis of binding of Na+, K+ and Li+ to GIPCs (wild-type) and IPCs (moca1).
a, b, ITC analysis of binding of Na+ to IPCs. Lipid vesicles were produced from the moca1 GIPC–IPC mixture as in Fig. 5b with >90% IPCs (Fig. 5d). The ITC data (a) and the plots of injected heat for automatic injections of NaCl solution into the sample cell containing the vesicle solution (b) are shown. Six independent ITC experiments were performed with similar results. c–f, The number of sites (c), heat change (ΔH) (d), entropy change (ΔS) (e), and binding constants (f) were derived from the fitted ITC from experiments similar to those in Fig. 6a, b for wild-type (GIPCs) and those in a, b for moca1 (IPCs). Mean ± s.d., n = 6; ***P < 0.001; NS, not significant; d, P = 0.787; e, P = 0.341; f, P = 0.407). g, h, ITC analysis of binding of K+ to GIPCs (wild-type) and IPCs (moca1). The number of sites (g) and binding constants (h) were derived from the fitted ITC from experiments similar to those for Na+ binding to GIPCs and IPCs. Mean ± s.d., n = 8; **P = 0.017; NS, not significant; h, P = 0.442). The number of apparent K+-binding sites for GIPCs and IPCs was 2.85 ± 0.35 and 2.42 ± 0.28, respectively (P < 0.01); Kd was 1.873 ± 0.823 mM and 1.338 ± 0.190 mM, respectively (P > 0.05). i, j, ITC analysis of binding of Li+ to GIPCs (wild-type) and IPCs (moca1). The number of sites (i) and binding constants (j) were derived from the fitted ITC. Mean ± s.d., n = 8; ***P < 0.001; NS, not significant; j, P = 0.878). The number of apparent Li+-binding sites for GIPCs and IPCs was 3.36 ± 0.18 and 2.93 ± 0.16, respectively (P < 0.001); Kd was 2.857 ± 1.273 mM and 2.873 ± 1.209 mM, respectively (P > 0.05). The Kd values for binding of the three monovalent cations to GIPCs are comparable to the estimated Kd for Na+ in depolarizing ζ potentials (Fig. 5e), but much smaller than the estimated Kd for these cations in triggering increases in [Ca2+]i (Figs. 1a, 3d–f), largely owing to the dilution of these cations by apoplastic water when the cations go through stomatal pores to reach the apoplastic side of the plasma membrane.
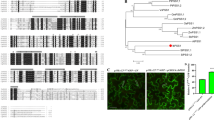
Extended Data Fig. 10 Phylogeny of GT8 domains in fully sequenced eukaryotic and bacterial genomes.
UTP-glucose-1-phosphate uridylyltransferase (UDPGP) domain-containing proteins in humans, yeast, and Arabidopsis were selected as outgroups to root the gene tree. UDPGP and GT8 belong to the same pfam clan GT_A (CL0110). Six groups of Arabidopsis GT8 genes, three groups of metazoan genes, and another three groups of fungal genes are identified and highlighted. Domain structures of plant GT8 genes are labelled similarly to these in Fig. 4a. The UDPGP domain-containing proteins are NP_006750, XP_005245519, and XP_006717380 in humans; NP_012889, NP_011851, and NP_010180.1 in yeast; and NP_197233, NP_186975, NP_181047, and NP_564372 in Arabidopsis.
Supplementary information
41586_2019_1449_MOESM2_ESM.avi
Video 1 NaCl-induced [Ca2+]i wave-like propagation in wild-type and moca1 roots. Transmission of Ca2+ wave through tissue shootward of the root tip (down) locally treated with 200 mM NaCl in wild-type and moca1, which were placed side-by-side. The experiments were performed as described in Fig. 3g. [Ca2+]i was monitored using Arabidopsis seedlings expressing the YC3.6 Ca2+ indicator and fluorescence ratio imaging. Video duration was 180 s (2 s per frame, 90 frames).
Source data
Rights and permissions
About this article
Cite this article
Jiang, Z., Zhou, X., Tao, M. et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 572, 341–346 (2019). https://doi.org/10.1038/s41586-019-1449-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1449-z
This article is cited by
-
Integrative physiology and transcriptome reveal salt-tolerance differences between two licorice species: Ion transport, Casparian strip formation and flavonoids biosynthesis
BMC Plant Biology (2024)
-
A positive role of polyunsaturated fatty acids on sustainable crop production against salt stress: an overview
Biologia (2024)
-
Exploring the plant lipidome: techniques, challenges, and prospects
Advanced Biotechnology (2024)
-
Science fosters ongoing reassessments of plant capabilities
Theoretical and Experimental Plant Physiology (2024)
-
Characters of the MOCA family in wheat and TaMOCA1 function in salt stress tolerance
Journal of Genetics (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.