Abstract
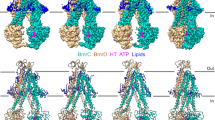
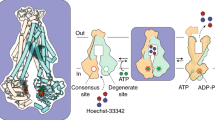
Cryo-electron microscopy (cryo-EM) has the capacity to capture molecular machines in action1,2,3. ATP-binding cassette (ABC) exporters are highly dynamic membrane proteins that extrude a wide range of substances from the cytosol4,5,6 and thereby contribute to essential cellular processes, adaptive immunity and multidrug resistance7,8. Despite their importance, the coupling of nucleotide binding, hydrolysis and release to the conformational dynamics of these proteins remains poorly resolved, especially for heterodimeric and/or asymmetric ABC exporters that are abundant in humans. Here we present eight high-resolution cryo-EM structures that delineate the full functional cycle of an asymmetric ABC exporter in a lipid environment. Cryo-EM analysis under active turnover conditions reveals distinct inward-facing (IF) conformations—one of them with a bound peptide substrate—and previously undescribed asymmetric post-hydrolysis states with dimerized nucleotide-binding domains and a closed extracellular gate. By decreasing the rate of ATP hydrolysis, we could capture an outward-facing (OF) open conformation—an otherwise transient state vulnerable to substrate re-entry. The ATP-bound pre-hydrolysis and vanadate-trapped states are conformationally equivalent; both comprise co-existing OF conformations with open and closed extracellular gates. By contrast, the post-hydrolysis states from the turnover experiment exhibit asymmetric ATP and ADP occlusion after phosphate release from the canonical site and display a progressive separation of the nucleotide-binding domains and unlocking of the intracellular gate. Our findings reveal that phosphate release, not ATP hydrolysis, triggers the return of the exporter to the IF conformation. By mapping the conformational landscape during active turnover, aided by mutational and chemical modulation of kinetic rates to trap the key intermediates, we resolved fundamental steps of the substrate translocation cycle of asymmetric ABC transporters.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All nine three-dimensional cryo-EM density maps of the heterodimeric/asymmetric ABC transporter TmrAB in nanodiscs have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-4773, EMD-4774, EMD-4775, EMD-4776, EMD-4777, EMD-4778, EMD-4779, EMD-4780 and EMD-4781 (see Extended Data Table 1). Atomic coordinates for the atomic models have been deposited in the Protein Data Bank under accession numbers 6RAF, 6RAG, 6RAH, 6RAI, 6RAJ, 6RAK, 6RAL, 6RAM and 6RAN. All other data are available from the corresponding authors upon reasonable request.
References
Henderson, R. Realizing the potential of electron cryo-microscopy. Q. Rev. Biophys. 37, 3–13 (2004).
Frank, J. New opportunities created by single-particle cryo-EM: the mapping of conformational space. Biochemistry 57, 888–888 (2018).
Cheng, Y. Single-particle cryo-EM—how did it get here and where will it go. Science 361, 876–880 (2018).
Rees, D. C., Johnson, E. & Lewinson, O. ABC transporters: the power to change. Nat. Rev. Mol. Cell Biol. 10, 218–227 (2009).
Locher, K. P. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487–493 (2016).
Thomas, C. & Tampé, R. Multifaceted structures and mechanisms of ABC transport systems in health and disease. Curr. Opin. Struct. Biol. 51, 116–128 (2018).
Robey, R. W. et al. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer 18, 452–464 (2018).
Trowitzsch, S. & Tampé, R. ABC transporters in dynamic macromolecular assemblies. J. Mol. Biol. 430, 4481–4495 (2018).
Dean, M. & Annilo, T. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu. Rev. Genomics Hum. Genet. 6, 123–142 (2005).
Zutz, A. et al. Asymmetric ATP hydrolysis cycle of the heterodimeric multidrug ABC transport complex TmrAB from Thermus thermophilus. J. Biol. Chem. 286, 7104–7115 (2011).
Kim, J. et al. Subnanometre-resolution electron cryomicroscopy structure of a heterodimeric ABC exporter. Nature 517, 396–400 (2015).
Nöll, A. et al. Crystal structure and mechanistic basis of a functional homolog of the antigen transporter TAP. Proc. Natl Acad. Sci. USA 114, E438–E447 (2017).
Alam, A., Kowal, J., Broude, E., Roninson, I. & Locher, K. P. Structural insight into substrate and inhibitor discrimination by human P-glycoprotein. Science 363, 753–756 (2019).
Dey, S., Ramachandra, M., Pastan, I., Gottesman, M. M. & Ambudkar, S. V. Evidence for two nonidentical drug-interaction sites in the human P-glycoprotein. Proc. Natl Acad. Sci. USA 94, 10594–10599 (1997).
Grossmann, N. et al. Mechanistic determinants of the directionality and energetics of active export by a heterodimeric ABC transporter. Nat. Commun. 5, 5419 (2014).
Choudhury, H. G. et al. Structure of an antibacterial peptide ATP-binding cassette transporter in a novel outward occluded state. Proc. Natl Acad. Sci. USA 111, 9145–9150 (2014).
Dawson, R. J. & Locher, K. P. Structure of a bacterial multidrug ABC transporter. Nature 443, 180–185 (2006).
Dawson, R. J. & Locher, K. P. Structure of the multidrug ABC transporter Sav1866 from Staphylococcus aureus in complex with AMP-PNP. FEBS Lett. 581, 935–938 (2007).
Zaitseva, J. et al. A structural analysis of asymmetry required for catalytic activity of an ABC-ATPase domain dimer. EMBO J. 25, 3432–3443 (2006).
Oldham, M. L. & Chen, J. Snapshots of the maltose transporter during ATP hydrolysis. Proc. Natl Acad. Sci. USA 108, 15152–15156 (2011).
Yang, M. et al. Single-molecule probing of the conformational homogeneity of the ABC transporter BtuCD. Nat. Chem. Biol. 14, 715–722 (2018).
Verhalen, B. et al. Energy transduction and alternating access of the mammalian ABC transporter P-glycoprotein. Nature 543, 738–741 (2017).
Watanabe, R., Iino, R. & Noji, H. Phosphate release in F1-ATPase catalytic cycle follows ADP release. Nat. Chem. Biol. 6, 814–820 (2010).
Okazaki, K. & Hummer, G. Phosphate release coupled to rotary motion of F1-ATPase. Proc. Natl Acad. Sci. USA 110, 16468–16473 (2013).
Catipovic, M. A., Bauer, B. W., Loparo, J. J. & Rapoport, T. A. Protein translocation by the SecA ATPase occurs by a power-stroke mechanism. EMBO J. 38, e101140 (2019).
Roos, C. et al. Characterization of co-translationally formed nanodisc complexes with small multidrug transporters, proteorhodopsin and with the E. coli MraY translocase. Biochim. Biophys. Acta 1818, 3098–3106 (2012).
Ehrnstorfer, I. A., Geertsma, E. R., Pardon, E., Steyaert, J. & Dutzler, R. Crystal structure of a SLC11 (NRAMP) transporter reveals the basis for transition-metal ion transport. Nat. Struct. Mol. Biol. 21, 990–996 (2014).
Pardon, E. et al. A general protocol for the generation of nanobodies for structural biology. Nat. Protoc. 9, 674–693 (2014).
Zimmermann, I. et al. Synthetic single domain antibodies for the conformational trapping of membrane proteins. eLife 7, e34317 (2018).
Gewering, T., Januliene, D., Ries, A. B. & Moeller, A. Know your detergents: A case study on detergent background in negative stain electron microscopy. J. Struct. Biol. 203, 242–246 (2018).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
Tegunov, D. & Cramer, P. Real-time cryo-EM data pre-processing with Warp. Preprint at https://www.biorxiv.org/content/10.1101/338558v1 (2018).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Šali, A. & Blundell, T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993).
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005).
Singharoy, A. et al. Molecular dynamics-based refinement and validation for sub-5 Å cryo-electron microscopy maps. eLife 5, e16105 (2016).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thronton, J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Wu, E. L. et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 35, 1997–2004 (2014).
Best, R. B. et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012).
Abraham, M. J. et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015).
Han, W., Cheng, R. C., Maduke, M. C. & Tajkhorshid, E. Water access points and hydration pathways in CLC H+/Cl− transporters. Proc. Natl Acad. Sci. USA 111, 1819–1824 (2014).
Khelashvili, G. et al. Conformational dynamics on the extracellular side of LeuT controlled by Na+ and K+ ions and the protonation state of Glu290. J. Biol. Chem. 291, 19786–19799 (2016).
Zhao, C. & Noskov, S. Y. The role of local hydration and hydrogen-bonding dynamics in ion and solute release from ion-coupled secondary transporters. Biochemistry 50, 1848–1856 (2011).
Lin, D. Y., Huang, S. & Chen, J. Crystal structures of a polypeptide processing and secretion transporter. Nature 523, 425–430 (2015).
Kim, Y. & Chen, J. Molecular structure of human P-glycoprotein in the ATP-bound, outward-facing conformation. Science 359, 915–919 (2018).
Mi, W. et al. Structural basis of MsbA-mediated lipopolysaccharide transport. Nature 549, 233–237 (2017).
Perez, C. et al. Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature 524, 433–438 (2015).
Johnson, Z. L. & Chen, J. ATP binding enables substrate release from multidrug resistance protein 1. Cell 172, 81–89.e10 (2018).
Hohl, M. et al. Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter. Proc. Natl Acad. Sci. USA 111, 11025–11030 (2014).
Acknowledgements
This research was supported by the German Research Foundation (Mo2752/2 to A.M., SFB 807 – Membrane Transport and Communication to E.R.G., G.H., A.M. and R.T., and Cluster of Excellence Frankfurt EXC 115 – Macromolecular Complexes to E.R.G., G.H., A.M. and R.T.). D.J., A.R.M., G.H. and A.M. acknowledge support from the Max Planck Society. R.T. is grateful for the support from an ERC Advanced Grant (789121). We acknowledge W. Kühlbrandt and D. Mills for access to the cryo-EM facility at the Max Planck Institute of Biophysics funded by the Max Planck Society; K. Holzhüter for creating the nanobody libraries; and S. Štefanić from the Nanobody Service Facility (University of Zurich) for alpaca immunization and lymphocyte isolation. We thank all members of the Institute of Biochemistry (Goethe University Frankfurt) and the staff at the Department of Structural Biology (Max Planck Institute of Biophysics) for discussions and comments on the manuscript.
Author information
Authors and Affiliations
Contributions
S.H. prepared all TmrAB samples and performed nucleotide occlusion, ATP hydrolysis, and transport experiments. S.H. and D.J. performed the negative-stain electron microscopy analyses. D.J. and A.M. carried out all cryo-EM imaging and single-particle analyses. A.R.M. and G.H. performed and analysed the molecular dynamics simulations. A.R.M. and C.T. built the structural models. B.T.K. and E.R.G. generated and selected nanobodies, and performed their characterization together with S.B., E.S. and S.H. S.H. and S.B. generated, purified and characterized all mutants. S.H., D.J., A.R.M., G.H., R.T. and A.M. interpreted the data and wrote the manuscript with contributions from all authors. R.T. and A.M. conceived the study, designed the research and planned the experiments. R.T. initiated, planned and coordinated the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peer review information Nature thanks Sergei Noskov, John Rubinstein and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Fig. 1 Reconstitution of TmrAB in lipid nanodiscs.
a, SEC and SDS–PAGE (Coomassie) illustrating reconstitution of TmrAB in MSP1D1 nanodiscs. A Superdex 200 Increase 3.2/300 column was used. b, Isolation of the TmrAB–MSP–Nb complex for cryo-EM. A Shodex KW404-4F column was used. c, Time course of [γ-32P]ATP hydrolysis of detergent-solubilized and nanodisc-reconstituted wild-type TmrAB (TmrABWT) and TmrAEQB. d, ATPase activity of detergent-solubilized and nanodisc-embedded wild-type TmrAB and TmrAEQB (background corrected). TmrAEQB displays a markedly decreased ATPase activity in comparison to wild-type TmrAB. The mean ± s.d. is calculated on the basis of three different data points (time points in Fig. 1c).
Extended Data Fig. 2 TmrAB–nanobody interaction.
a, Expanded view of a representative negative-stain raw micrograph and 2D class averages of TmrAB–Nb complexes. In the 2D class averages, note the clear density for Nb9F10 bound to the intracellular side of TmrAB. The scale bar is 100 nm in the micrograph and 25 nm in the inset (magnified 4×). b, Peptide transport of TmrAB in proteoliposomes with and without addition of Nb9F10. Proteoliposomes were incubated with Nb9F10 (100 nM) for 15 min on ice and peptide import into proteoliposomes was subsequently followed for 8 min at 45 °C; mean ± s.d. from three experiments. c, d, Nb9F10 binds the apo (c) and vanadate-trapped (d) TmrAB. Data are normalized to apo TmrAB without Nb. A TSKgel G3000SWXL column was used. e, f, Nb9F10 forms a stable complex with NBD of TmrB (f), but not TmrA (e). To illustrate stoichiometric binding, data were normalized to the NBDTmrA chromatogram without Nb. A Superdex 200 Increase 3.2/300 column was used. g, Surface representation of TmrAB and Nb9F10, coloured as in Fig. 1. CDR2 and CDR3 are shown in orange and red, respectively. h, Close-up view of the TmrAB/Nb9F10 interface, coloured as in g. Polar interactions include a short β-sheet between the CDR3 loop and a β-strand of the NBD as well as several residues of CDR2, which are shown as sticks.
Extended Data Fig. 3 Cryo-EM densities and conformational distribution from all datasets.
a, Final maps from all datasets. In all cases, at least two conformations were observed, in accordance with the highly dynamic nature of the transporter. b, Particle distribution in each dataset. The first column represents the total number of particles after two rounds of 3D classification to remove false positives. At this stage, the main OF and IF conformations were already distinguishable (IF particles, orange; OF, blue; UR, teal; see also Extended Data Fig. 5); however, no safe assignment of the sub-conformation could be made. The second column represents the distribution of each specific conformation after further sub-classification. Colour code as in a (the two URasym conformations are coloured in light and dark green, poor-quality particles are grey). c, FSC curves for all maps in each dataset, along with the number of particles used during the final refinement.
Extended Data Fig. 4 Cryo-EM analysis of TmrAEQBATP-ADP.
a, Typical cryo-EM micrograph. Scale bars are 50 nm in the micrograph and 20 nm in the magnified view. b, Representative 2D class averages for each conformation observed in the dataset. c, FSC curves, generated in RELION-3, for all TmrAEQBATP-ADP maps. d, Angular assignment for each map. The height of individual cylinder bars is proportional to the number of particles in each view. The most frequent views are coloured in red. e, Local-resolution estimation for the four densities as estimated in CryoSPARC.
Extended Data Fig. 5 Image-processing workflow for TmrABturnover.
From 7,266 original motion-corrected movies, 6,937 micrographs—containing a signal better than 5 Å resolution (with the majority around 3.5 Å) as estimated by Gctf35—were used in processing. Four million particles were generously auto-picked in RELION-3 using 2D templates, extracted at a box size of 64 pixels with 4.3 Å per pixel and directly subjected to multi-model 3D classification to eliminate false positives. To speed up the calculations and overcome large memory demands, the stack was divided into two subsets—containing two million particles each—which were processed in parallel. All maps that reached 8.6 Å resolution (Nyquist for the binned data) were kept (orange) and the corresponding particles were re-centred, re-extracted at full pixel size and subjected to a second round of 3D classification (rejected maps are shown in grey). The selected IF (orange) and URasym (blue) particles were further subclassified. The resulting IFwide (light yellow), IFnarrow (yellow) and URasym (blue) particles were pooled and each conformation was refined separately. Particles were polished and ctf-refined as implemented in RELION-3, resulting in 4.4 Å, 4.1 Å and 3.6 Å maps, respectively. Despite the overall high quality of the obtained densities, visual inspection still revealed considerable anisotropy in the resulting volumes. We interpreted this as the result of remaining conformational variance and therefore re-classified the individual stacks to sort out conformational flexibility. To do this, the refined URasym particles were subjected to a fourth round of 3D classification with no alignment, which revealed two different URasym conformations. Both conformations were further refined using non-uniform refinement in CryoSPARC36, resulting in 3.5 Å and 3.8 Å resolution maps for URasym (green) and URasym∗ (light green), respectively. In parallel, IF particles were subjected to heterogenous refinement, followed by non-uniform refinement in CryoSPARC, which further improved the resolution to 3.8 and 4.2 Å for IFnarrow and IFwide, respectively. Colour code as in Extended Data Fig. 3. Processing workflows for all other datasets followed the same scheme with only minor modifications at the last steps of 3D classification.
Extended Data Fig. 6 Map quality.
To demonstrate the quality of each map, a selected region—spanning residues 213–235 of TmrA and 94–106 of TmrB—was extracted from each corresponding experimental map. All maps display clear side-chain densities, except for the IFwide map of the TmrABapo control, which is also the smallest dataset in size.
Extended Data Fig. 7 Conformation space of type I ABC exporters.
a–c, Conformation space of the cryo-EM structures from this study and previously published ABC tranporter structures is shown based on three parameters: the intracellular gate angle (IC angle), the extracellular gate angle (EC angle), and the distance between two nucleotide-binding domains (NBD distance). The electron microscopy structures obtained in this study are shown as blue circles, and previously published structures are shown as orange triangles. The clustering of structures in three major conformations (IF, OFopen and OFoccluded) is highlighted by dashed rectangles in a and b. The cryo-EM structures obtained in this study cover all main conformations. The new URasym conformations show the closest resemblance to OFoccluded. The IC angle is defined as the angle between two vectors: vector 1 between the centre of mass of the whole extracellular part of the TMD region and the centre of mass of the intracellular parts of TM1TmrA, TM2TmrA, TM3TmrA, TM6TmrA, TM4TmrB and TM5TmrB; and vector 2 between the centre of mass of the whole extracellular part of the TMD region and the centre of mass of the intracellular parts of TM1TmrB, TM2TmrB, TM3TmrB, TM6TmrB, TM4TmrA and TM5TmrA. The EC angle is defined as the angle between two vectors: vector 3 between the centres of mass of the NBDs and the extracellular parts TM1TrmA, TM2TrmA, TM3TrmB, TM4TrmB, TM5TrmB and TM6TrmB; and vector 4 between the centre of mass of the NBDs and of the extracellular parts TM1TmrB, TM2TmrB, TM3TmrA, TM4TmrA, TM5TmrA and TM6TmrA. The NBD distance is between the centres of mass of two NBDs (N-terminal loops and C-terminal helices were excluded). IF, IF conformation; O, OFopen conformation; C, OFoccluded conformation; Vi, TmrABATP-Vi; EQ, TmrAEQB. PDB IDs: PCAT, 4RY2 and 4S0F for the IF and OFoccluded conformations, respectively51; McjD, 4PL016; Pgp, 6C0V52; MsbA, 5TTP53; PglK, 5C7354; MRP, 6BHU55; Sav, 2ONJ18; TmrAB, 5MKK12; TM287 (TM287/288), 4Q4A56. d, Time-dependent IC angle in independent molecular dynamics simulations of IFwide and IFnarrow, with ATP/ATP, ADP/ATP and ADP/ADP in the canonical/non-canonical sites, respectively. Horizontal lines indicate the IC angles of the IFnarrow and IFwide structures.
Supplementary information
Supplementary Figure 1
This file contains the gel source data and uncropped blots.
Video 1: Conformational space of TmrAB
Video representation of the distinct conformations of TmrAB, as determined by cryo-EM. The IF confromations display different degrees of intracellular gate opening. ATP-bound and vanadate-trapped states are equivalent and co-exist in OFopen and OFoccluded conformations. The asymmetric URasym and URasym∗ conformations display a progressive opening of the intracellular gate and represent a post-hydrolysis state on path to reset the transporter to the IF conformation. Data is represented as EM-densities and corresponding molelucar models. TmrA is colored in blue, TmrB in yellow and the nanobody in light grey.
Rights and permissions
About this article
Cite this article
Hofmann, S., Januliene, D., Mehdipour, A.R. et al. Conformation space of a heterodimeric ABC exporter under turnover conditions. Nature 571, 580–583 (2019). https://doi.org/10.1038/s41586-019-1391-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1391-0
This article is cited by
-
Structural basis of prostaglandin efflux by MRP4
Nature Structural & Molecular Biology (2024)
-
Expulsion mechanism of the substrate-translocating subunit in ECF transporters
Nature Communications (2023)
-
Structural remodelling of the carbon–phosphorus lyase machinery by a dual ABC ATPase
Nature Communications (2023)
-
On the interplay between lipids and asymmetric dynamics of an NBS degenerate ABC transporter
Communications Biology (2023)
-
Asymmetric conformations and lipid interactions shape the ATP-coupled cycle of a heterodimeric ABC transporter
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.