Abstract
Activated CD4 T cells proliferate rapidly and remodel epigenetically before exiting the cell cycle and engaging acquired effector functions. Metabolic reprogramming from the naive state is required throughout these phases of activation1. In CD4 T cells, T-cell-receptor ligation—along with co-stimulatory and cytokine signals—induces a glycolytic anabolic program that is required for biomass generation, rapid proliferation and effector function2. CD4 T cell differentiation (proliferation and epigenetic remodelling) and function are orchestrated coordinately by signal transduction and transcriptional remodelling. However, it remains unclear whether these processes are regulated independently of one another by cellular biochemical composition. Here we demonstrate that distinct modes of mitochondrial metabolism support differentiation and effector functions of mouse T helper 1 (TH1) cells by biochemically uncoupling these two processes. We find that the tricarboxylic acid cycle is required for the terminal effector function of TH1 cells through succinate dehydrogenase (complex II), but that the activity of succinate dehydrogenase suppresses TH1 cell proliferation and histone acetylation. By contrast, we show that complex I of the electron transport chain, the malate–aspartate shuttle and mitochondrial citrate export are required to maintain synthesis of aspartate, which is necessary for the proliferation of T helper cells. Furthermore, we find that mitochondrial citrate export and the malate–aspartate shuttle promote histone acetylation, and specifically regulate the expression of genes involved in T cell activation. Combining genetic, pharmacological and metabolomics approaches, we demonstrate that the differentiation and terminal effector functions of T helper cells are biochemically uncoupled. These findings support a model in which the malate–aspartate shuttle, mitochondrial citrate export and complex I supply the substrates needed for proliferation and epigenetic remodelling early during T cell activation, whereas complex II consumes the substrates of these pathways, which antagonizes differentiation and enforces terminal effector function. Our data suggest that transcriptional programming acts together with a parallel biochemical network to enforce cell state.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request. RNA-seq datasets have been deposited in Gene Expression Omnibus under the accession number GSE130713.
References
Buck, M. D., Sowell, R. T., Kaech, S. M. & Pearce, E. L. Metabolic instruction of immunity. Cell 169, 570–586 (2017).
Buck, M. D., O’Sullivan, D. & Pearce, E. L. T cell metabolism drives immunity. J. Exp. Med. 212, 1345–1360 (2015).
Klein Geltink, R. I. et al. Mitochondrial priming by CD28. Cell 171, 385–397.e11 (2017).
Buck, M. D. et al. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 166, 63–76 (2016).
Chang, C.-H. et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251 (2013).
Wang, R. et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882 (2011).
Peng, M. et al. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science 354, 481–484 (2016).
Peters, R. Biochemical Lesions and Lethal Synthesis (Pergamon, 1963).
Contreras, L. & Satrústegui, J. Calcium signaling in brain mitochondria: interplay of malate aspartate NADH shuttle and calcium uniporter/mitochondrial dehydrogenase pathways. J. Biol. Chem. 284, 7091–7099 (2009).
Safer, B. The metabolic significance of the malate-aspartate cycle in heart. Circ. Res. 37, 527–533 (1975).
LaNoue, K. F. & Williamson, J. R. Interrelationships between malate-aspartate shuttle and citric acid cycle in rat heart mitochondria. Metabolism 20, 119–140 (1971).
Wellen, K. E. et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 (2009).
Birsoy, K. et al. An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 162, 540–551 (2015).
Sullivan, L. B. et al. Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell 162, 552–563 (2015).
Platt, R. J. et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014).
Gagliani, N. et al. Coexpression of CD49b and LAG-3 identifies human and mouse T regulatory type 1 cells. Nat. Med. 19, 739–746 (2013).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Anders, S., Py, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Lu, W., Wang, L., Chen, L., Hui, S. & Rabinowitz, J. D. Extraction and quantitation of nicotinamide adenine dinucleotide redox cofactors. Antioxid. Redox Signal. 28, 167–179 (2018).
Jang, C. et al. The small intestine converts dietary fructose into glucose and organic acids. Cell Metab. 27, 351–361.e3 (2018).
Melamud, E., Vastag, L. & Rabinowitz, J. D. Metabolomic analysis and visualization engine for LC-MS data. Anal. Chem. 82, 9818–9826 (2010).
Acknowledgements
This work was supported by NIH grants R37 AR40072, R61AR073048 (J.C. and R.A.F.), F31 AI1333855 (J.A.S.), T32 AI7019-41 (J.A.S.), R01 CA166025-04 (L.J.M. III), T32 GM065841-14 (L.J.M. III), the Howard Hughes Medical Institute (R.A.F.), European Union’s Horizon 2020, and Marie Sklodowska-Curie grant agreement no. 751423 (J.C.G.C.), and the Paradifference Foundation (L.J.M. III).
Peer review information
Nature thanks Navdeep Chandel and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
W.B., J.A.S., J.C. and R.A.F. designed the study and wrote the manuscript. W.B. and J.A.S. designed and performed experiments. J.Z., R.Q. and Y.K. performed all bioinformatic and genomic analysis. P.B. assisted with sequencing. J.C.G.C. and J.R. designed and performed LC–MS experiments and data analysis. F.J.A.K. and L.J.M. III prepared and provided Sdhc cKO mouse tissue. O.K. assisted with vector cloning. H.R.S. assisted with experimentation. R.J. assisted with experimental design. All authors edited and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
R.A.F. is a founder, shareholder and advisor for Rheos Medicines, Inc.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
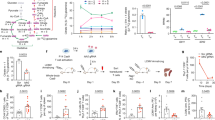
Extended Data Fig. 1 Acute ETC inhibition alters cell cycle, but not viability, in TH1 cells.
a, Viability measured by propidium iodide (PI) and annexin-V staining of wild-type CD4 T cells cultured in TH1 conditions and treated overnight for 16 h on day 1, 2 or 3 of culture with DMSO, rotenone, DMM, antimycin A or oligomycin (n = 3). b, Cell-cycle analysis measured by Ki67 and DAPI of CD4 T cells cultured in TH1 conditions on day 3 following 16-h overnight treatment with DMSO (n = 5), rotenone, DMM, antimycin A or oligomycin (n = 6). n, number of technical replicates. Representative plots and a graph summarizing the results of three independent experiments are shown. Mean and s.d. of replicates are presented on summarized plots and unpaired, two-tailed t-test used to determine significance. *P < 0.05, **P < 0.01.
Extended Data Fig. 2 ETC regulation of proliferation is conserved among subtypes of T helper cells, but ETC requirements for effector cytokine transcription differ between TH1, TH2 and TH17 cells.
a, b, Proliferation of wild-type CD4 T cells cultured in TH2 (a) and TH17 (b) conditions, following 16-h overnight treatment with DMSO, rotenone or oligomycin (n = 3). c, d, Cell-cycle analysis measured by Ki67 and DAPI of CD4 T cells cultured in TH2 (c) and TH17 (d) conditions on day 3 following 16-h overnight treatment with DMSO, rotenone, DMM, antimycin A or oligomycin (n = 6). e, f, Effector cytokine transcription after PMA and ionomycin restimulation at day 5 measured by IL-4–GFP (4GET) reporter expression in cells cultured in TH2 conditions (e) and IL17–GFP reporter expression in cells cultured in TH17 conditions (f) following 16-h overnight treatment with DMSO, rotenone, DMM, antimycin A or oligomycin (n = 3). n, number of technical replicates. Representative plots and a graph summarizing the results of three independent experiments are shown. Mean and s.d. of replicates are presented on summarized plots and unpaired, two-tailed t-test used to determine significance. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Fig. 3 Complex II inhibition is functional and leads to a loss of IFNγ production in TH1 cells.
a, Cellular succinate at day 5 evaluated using Succinate Assay Kit (Abcam) in wild-type CD4 T cells cultured in TH1 conditions following 6-h treatment with DMSO, 10 mM DMM, 1 mM 3NP, 100 μM TTFA or 1 μM atpenin A5 (n = 3). b, IFNγ protein production after PMA and ionomycin restimulation at day 5 of wild-type CD4 T cells cultured in TH1 conditions following 16-h overnight treatment with 10 mM diethyl succinate (DES) (n = 5) or DMSO (n = 4). c, IFNγ protein production after PMA and ionomycin restimulation at day 5 of Cas9-expressing CD4 T cells cultured in TH1 conditions transduced with one of three individual sgRNA targeting Sdha, or an empty-vector control (n = 3 biological replicates). d, e, Total cellular succinate (d) and α-ketoglutarate (e) measured by LC–MS analysis in wild-type or Sdhc cKO CD4 T cells cultured in TH1 conditions after 4-h culture in dialysed FBS-containing medium at day 5 (n = 3). n, number of technical replicates, unless otherwise stated. Representative plots and a graph summarizing the results of at least two independent experiments are shown. Mean and s.d. of replicates are presented on summarized plots and unpaired, two-tailed t-test used to determine significance. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Fig. 4 Retroviral expression of sgRNA in Cas9-expressing CD4 T cells.
a, Schematic of MG-Guide retroviral vector. b, CD4 T cells from Cas9-expressing mice were stimulated with anti-CD3 and anti-CD28 coated beads for 24 h and retrovirally transduced with either a MG-Guide (empty vector) or a MG-Guide vector cloned to express a sgRNA against Thy1 (Thy1 sgRNA). GFP expression was measured at 24 h after transduction, compared to non-transduced cells. c, THY1.1 protein expression was measured in transduced (empty vector blue line; Thy1 sgRNA red line) and non-transduced (solid grey) cells by flow cytometry at 30 and 96 h after transduction. d, Schematic of experimental design for functional TH1 sgRNA studies. e, CD4 T cells from Cas9-expressing mice were stimulated with anti-CD3 and anti-CD28 beads in IL-2 (5 ng ml−1), anti-IL-4 (10 μg ml−1) and IL-12 (2 ng ml−1) and retrovirally transduced 24 h after activation with either empty MG-Guide (shaded blue) or MG-Guide expressing a sgRNA against Tbx21 (outline). TBET protein expression was measured by intracellular flow cytometry on day 3. f, Cas9-expressing CD4 T cells were cultured as above, and infected with MG-Guide, a sgRNA against Tbx21 or a sgRNA against Il12rb. IFNγ protein was measured by intracellular flow cytometry on day 5 after restimulation with PMA (20 ng ml−1) and ionomycin (1 μg ml−1).
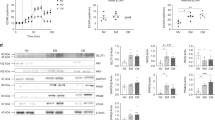
Extended Data Fig. 5 Complex II regulates epigenetic modifications and program-specific gene expression in TH1 cells.
a, H3K9 acetylation and H3K27 acetylation, normalized to total cellular H3 and 1× DNA content on day 3 of wild-type CD4 T cells cultured in TH1 conditions after 16-h overnight treatment with DMSO, DMM, oligomycin, BMS-303141 or butyrate (n = 3). b, c, H3K9 acetylation (b) and H3K27 acetylation (c) at day 5 of Cas9-expressing CD4 T cells cultured in TH1 conditions transduced with one of three individual sgRNA targeting Sdha, or an empty-vector control (n = 3 biological replicates). d, e, Volcano plot summarizing RNA-seq data that indicate the most-differentially regulated transcripts between wild-type and Sdhc cKO TH1 cells (d) and gene set enrichment analysis (GSEA) plot of the GO cytokine production pathway (e) (n = 3 biological replicates). n, number of technical replicates unless otherwise stated. Representative plots and a graph summarizing the results of at least two independent experiments are shown. Mean and s.d. of replicates are presented on summarized plots and unpaired, two-tailed t-test used to determine significance. *P < 0.05, **P < 0.01.
Extended Data Fig. 6 The malate–aspartate shuttle and mitochondrial citrate export dynamically regulate histone acetylation and program-specific gene expression in TH1 cells.
a–d, H3K27 acetylation (a), total cellular H3 (b), H3K9 acetylation normalized to total cellular H3 and 1× DNA content (c) and H3K27 acetylation normalized to total cellular H3 and 1× DNA content (d) on day 4 of Cas9-expressing CD4 T cells transduced with three individual sgRNAs targeting Acly, Slc25a1, Mdh1, Slc25a11 or Slc1a3, or empty vector, cultured in TH1 conditions (n = 3 biological replicates). e, f, Volcano plot summarizing RNA-seq data that indicate the most-differentially regulated transcripts at day 5 in Cas9-expressing CD4 T cells cultured in TH1 conditions transduced with empty vector or one sgRNA targeting Slc25a1 (e) or Slc25a11 (f) (n = 2 biological replicates). Representative plots and a graph summarizing the results of at least two independent experiments are shown. Mean and s.d. of replicates are presented on summarized plots and unpaired, two-tailed t-test used to determine significance. *P < 0.05.
Extended Data Fig. 7 The malate–aspartate shuttle and mitochondrial citrate export are required for proliferation in TH1 cells.
Proliferation of Cas9-expressing CD4 T cells transduced with empty-vector control or one of three individual sgRNAs targeting Acly, Slc25a1, Mdh1, Slc25a11 or Slc1a3, cultured in TH1 conditions at day 5 (n = 3 biological replicates). Representative plots and a graph summarizing the results of at least two independent experiments are shown. Mean and s.d. of replicates are presented on summarized plots and unpaired, two-tailed t-test used to determine significance. *P < 0.05, **P < 0.01.
Extended Data Fig. 8 The malate–aspartate shuttle and mitochondrial citrate export regulate levels of cellular acetyl-CoA and cellular metabolism.
a, Cellular acetyl-CoA measured by LC–MS analysis in Cas9-expressing CD4 T cells transduced with empty vector or two individual sgRNAs targeting Slc25a1, as described, on day 5 of culture in TH1 conditions (n = 2 biological replicates, n = 3 technical replicates). a.u., arbitrary units. b, c, Volcano plot (b) and heat map (c) of all metabolites measured by LC–MS analysis in Cas9-expressing CD4 T cells transduced with empty vector or two individual sgRNAs targeting Slc25a1, as described, on day 5 of culture in TH1 conditions (n = 2 biological replicates, n = 3 technical replicates). d, e, Volcano plot (d) and heat map (e) of all metabolites measured by LC–MS analysis in Cas9-expressing CD4 T cells transduced with empty vector or two individual sgRNAs targeting Slc25a1, as described, on day 5 of culture in TH1 conditions (n = 2 biological replicates, n = 2 technical replicates). Mean and s.d. of replicates are presented on summarized plots and unpaired, two-tailed t-test used to determine significance. **P < 0.01, ****P < 0.0001.
Extended Data Fig. 9 Complex I activity is required for aspartate production and cell-cycle progression in activating TH1 cells.
a, b, Volcano plot (a) and heat map (b) of all metabolites measured by LC–MS analysis in wild-type CD4 T cells treated acutely for 4 h on day 5 of culture in TH1 conditions (n = 3). c, Cell-cycle analysis using Ki67 and DAPI of wild-type CD4 T cells cultured in TH1 conditions at day 3 following 16-h overnight treatment with DMSO or rotenone ± 20 mM aspartate (n = 3). n, number of technical replicates. Representative plots and a graph summarizing the results of at least two independent experiments are shown. Mean and s.d. of replicates are presented on summarized plots and unpaired, two-tailed t-test used to determine significance. **P < 0.01, ****P < 0.0001.
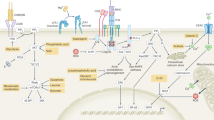
Extended Data Fig. 10 Conceptual models of mitochondrial metabolite transport and the consequence of metabolic perturbations on TH1 cell activation.
a, Early-stage TH1 cell activation is supported by the malate–aspartate shuttle and mitochondrial citrate export. These mitochondrial transport systems provide the key substrates that are needed for cell division and histone acetylation. Citrate export results in the production of cytosolic acetyl-CoA that can be used to synthesize the fatty acids that are needed for plasma membrane expansion during division, as well as the acetyl groups that are used for histone acetylation. Interconnected with this export pathway is the malate–aspartate shuttle, a carbon-neutral cycle that results in the net movement of NAD+ to the cytosol and NADH into the mitochondria, through which the cycle can fuel the activity of ETC complex I. Through the activity of complex I, NAD+ can be continually recycled, which enables the production of aspartate (an essential precursor for nucleotide synthesis). These processes are antagonized by the activity of SDH (ETC complex II), which consumes α-ketoglutarate; this limits the availability of the latter for the malate–aspartate shuttle and promotes effector functions of TH1 cells. b, T-helper-cell activation is defined by two major phases: (1) a period of rapid division and epigenetic remodelling, and (2) cell-cycle arrest and cytokine production. Each of these phases is supported by a discrete component of mitochondrial metabolism. The malate–aspartate shuttle and mitochondrial citrate export generate the material needed for early-phase cell differentiation to occur. As differentiation continues, the activity of complex II draws carbon away from the shuttle, and thus acts to pull activated TH1 cells out of the differentiation process and to enable them to fully engage their terminal effector cell program. When the mitochondrial transport networks are disrupted, TH1 cells are unable to properly proliferate or epigenetically reprogram. By contrast, inhibiting the activity of complex II causes activated TH1 cells to continuously proliferate and remodel their chromatin, which prevents them from exiting the differentiation phase and engaging their terminal effector program.
Supplementary information
Supplementary Table 1
sgRNA sequences used to target Thy1, Tbx21, Sdha, Acly, Slc25a1, Mdh2, Mdh1, Got1, Got2, Slc25a11, Slc1a3, and Cs using MG-Guide.
Supplementary Table 2
WT vs. Sdhc cKO RNA-seq significant genes p<0.05. RNA-seq of WT vs Sdhc cKO T cells at day 5 of activation in Th1 culture conditions.
Supplementary Table 3
EV vs. Slc25a1 sgRNA transduced cell RNA-seq significant genes p<0.05. RNA-seq of EV vs. Slc25a1 sgRNA transduced T cells at day 5 of activation in Th1 culture conditions.
Supplementary Table 4
EV vs. Slc25a11 sgRNA transduced cell RNA-seq significant genes p<0.05. RNA-seq of EV vs. Slc25a11 sgRNA transduced T cells at day 5 of activation in Th1 culture conditions.
Source data
Rights and permissions
About this article
Cite this article
Bailis, W., Shyer, J.A., Zhao, J. et al. Distinct modes of mitochondrial metabolism uncouple T cell differentiation and function. Nature 571, 403–407 (2019). https://doi.org/10.1038/s41586-019-1311-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1311-3
This article is cited by
-
Metabolic waypoints during T cell differentiation
Nature Immunology (2024)
-
Prognostic value of mitochondrial CKMT2 in Pan-cancer and its tumor immune correlation analysis
Scientific Reports (2024)
-
Amino acid metabolism in health and disease
Signal Transduction and Targeted Therapy (2023)
-
Metabolic regulation of somatic stem cells in vivo
Nature Reviews Molecular Cell Biology (2022)
-
Fasting as key tone for COVID immunity
Nature Metabolism (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.