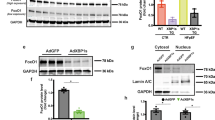
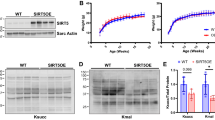
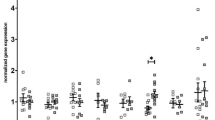
Abstract
Heart failure with preserved ejection fraction (HFpEF) is a common syndrome with high morbidity and mortality for which there are no evidence-based therapies. Here we report that concomitant metabolic and hypertensive stress in mice—elicited by a combination of high-fat diet and inhibition of constitutive nitric oxide synthase using Nω-nitro-l-arginine methyl ester (l-NAME)—recapitulates the numerous systemic and cardiovascular features of HFpEF in humans. Expression of one of the unfolded protein response effectors, the spliced form of X-box-binding protein 1 (XBP1s), was reduced in the myocardium of our rodent model and in humans with HFpEF. Mechanistically, the decrease in XBP1s resulted from increased activity of inducible nitric oxide synthase (iNOS) and S-nitrosylation of the endonuclease inositol-requiring protein 1α (IRE1α), culminating in defective XBP1 splicing. Pharmacological or genetic suppression of iNOS, or cardiomyocyte-restricted overexpression of XBP1s, each ameliorated the HFpEF phenotype. We report that iNOS-driven dysregulation of the IRE1α–XBP1 pathway is a crucial mechanism of cardiomyocyte dysfunction in HFpEF.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its Supplementary Information. Each data point corresponding to figures that describe the results from in vivo or in vitro model studies are provided as separate Source Data for Figs. 1c, d, f–i, 2a, d, f, g, i–m, 3a–d, f, g, i, j, 4a–d, f, h and Extended Data Figs. 1a–l, 2b–f, 3b–e, g, 4a–l, n, p, 5b–g, 6b–g, j–m, 7b, d, g, 8b–d, g, 9c–i, 10b–i. Other source data related to the study are available from the corresponding author upon reasonable request.
References
Dunlay, S. M., Roger, V. L. & Redfield, M. M. Epidemiology of heart failure with preserved ejection fraction. Nat. Rev. Cardiol. 14, 591–602 (2017).
Shah, S. J. et al. Phenotype-specific treatment of heart failure with preserved ejection fraction: a multiorgan roadmap. Circulation 134, 73–90 (2016).
Chirinos, J. A. & Zamani, P. The nitrate–nitrite–NO pathway and its implications for heart failure and preserved ejection fraction. Curr. Heart Fail. Rep. 13, 47–59 (2016).
Butler, J., Braunwald, E. & Gheorghiade, M. Recognizing worsening chronic heart failure as an entity and an end point in clinical trials. J. Am. Med. Assoc. 312, 789–790 (2014).
Roh, J., Houstis, N. & Rosenzweig, A. Why don’t we have proven treatments for HFpEF? Circ. Res. 120, 1243–1245 (2017).
Primessnig, U. et al. Novel pathomechanisms of cardiomyocyte dysfunction in a model of heart failure with preserved ejection fraction. Eur. J. Heart Fail. 18, 987–997 (2016).
Methawasin, M. et al. Experimentally increasing the compliance of titin through RNA binding motif-20 (RBM20) inhibition improves diastolic function in a mouse model of heart failure with preserved ejection fraction. Circulation 134, 1085–1099 (2016).
González-López, E. et al. Wild-type transthyretin amyloidosis as a cause of heart failure with preserved ejection fraction. Eur. Heart J. 36, 2585–2594 (2015).
Wang, Z. V. & Hill, J. A. Protein quality control and metabolism: bidirectional control in the heart. Cell Metab. 21, 215–226 (2015).
Wang, Z. V. et al. Spliced X-box binding protein 1 couples the unfolded protein response to hexosamine biosynthetic pathway. Cell 156, 1179–1192 (2014).
Rothermel, B. A. et al. Differential activation of stress-response signaling in load-induced cardiac hypertrophy and failure. Physiol. Genomics 23, 18–27 (2005).
Paulus, W. J. & Tschöpe, C. A novel paradigm for heart failure with preserved ejection fraction: comorbidities drive myocardial dysfunction and remodeling through coronary microvascular endothelial inflammation. J. Am. Coll. Cardiol. 62, 263–271 (2013).
Furfine, E. S., Harmon, M. F., Paith, J. E. & Garvey, E. P. Selective inhibition of constitutive nitric oxide synthase by l-N G-nitroarginine. Biochemistry 32, 8512–8517 (1993).
Kopincová, J., Púzserová, A. & Bernátová, I. l-NAME in the cardiovascular system — nitric oxide synthase activator? Pharmacol. Rep. 64, 511–520 (2012).
Yang, L. et al. S-Nitrosylation links obesity-associated inflammation to endoplasmic reticulum dysfunction. Science 349, 500–506 (2015).
Gould, N., Doulias, P. T., Tenopoulou, M., Raju, K. & Ischiropoulos, H. Regulation of protein function and signaling by reversible cysteine S-nitrosylation. J. Biol. Chem. 288, 26473–26479 (2013).
Moore, W. M. et al. l-N 6-(1-Iminoethyl)lysine: a selective inhibitor of inducible nitric oxide synthase. J. Med. Chem. 37, 3886–3888 (1994).
Borlaug, B. A., Koepp, K. E. & Melenovsky, V. Sodium nitrite improves exercise hemodynamics and ventricular performance in heart failure with preserved ejection fraction. J. Am. Coll. Cardiol. 66, 1672–1682 (2015).
Sharma, K. & Kass, D. A. Heart failure with preserved ejection fraction: mechanisms, clinical features, and therapies. Circ. Res. 115, 79–96 (2014).
Redfield, M. M. et al. isosorbide mononitrate in heart failure with preserved ejection fraction. N. Engl. J. Med. 373, 2314–2324 (2015).
Zamani, P. et al. Isosorbide dinitrate, with or without hydralazine, does not reduce wave reflections, left ventricular hypertrophy, or myocardial fibrosis in patients with heart failure with preserved ejection fraction. J. Am. Heart Assoc. 6, e004262 (2017).
Oeser, C. Heart failure: nitrates reduce activity levels in HFpEF. Nat. Rev. Cardiol. 13, 2–3 (2016).
Jia, J. et al. Target-selective protein S-nitrosylation by sequence motif recognition. Cell 159, 623–634 (2014).
Zhu, H. et al. Cardiac autophagy is a maladaptive response to hemodynamic stress. J. Clin. Invest. 117, 1782–1793 (2007).
Kong, Y. et al. Suppression of class I and II histone deacetylases blunts pressure-overload cardiac hypertrophy. Circulation 113, 2579–2588 (2006).
Oh, M. et al. Calcineurin is necessary for the maintenance but not embryonic development of slow muscle fibers. Mol. Cell. Biol. 25, 6629–6638 (2005).
Zhi, G. et al. Myosin light chain kinase and myosin phosphorylation effect frequency-dependent potentiation of skeletal muscle contraction. Proc. Natl Acad. Sci. USA 102, 17519–17524 (2005).
Wang, J. M. et al. IRE1α prevents hepatic steatosis by processing and promoting the degradation of select microRNAs. Sci. Signal. 11, eaao4617 (2018).
Acknowledgements
This work was supported by grants from NIH: HL-120732 (J.A.H.), HL-128215 (J.A.H.), HL-126012 (J.A.H.), F32HL136151 (K.M.F.), F32HL142244 (D.T.), 2T32HL007227-41 (V.S.H.), HL-135827 (D.A.K.), HL-119012 (D.A.K.), R01-HL102478 (P.P.A.M.), the American Heart Association (AHA): 16POST30680016 (F.A.), 16PRE29660003 (S.Y.K.), 14SFRN20510023 (J.A.H.), 14SFRN20670003 (J.A.H.) and 16SFRN28620000 (K.S. and D.A.K.), AHA and the Theodore and Beulah Beasley Foundation 18POST34060230 (G.G.S.), University Federico II of Naples and Compagnia di San Paolo STAR program (G.G.S.), Fondation Leducq TransAtlantic Network of Excellence11CVD04 (J.A.H.), Cancer Prevention and Research Institute of Texas RP110486P3 (J.A.H.), Senator Paul D. Wellstone Muscular Dystrophy Cooperative Research Center U54-HD087351 (P.P.A.M.) and by Fondo Nacional de Desarrollo Científico y Tecnológico, FONDAP 15130011 (S.L.). We thank G. S. Hotamisligil and L. Yang for providing ERN1 wild-type and ERN1 M1+M2-expressing adenoviruses. We thank A. Rosenberg for his help with the ProteinSimple capillary immunoassay analysis (partly supported by The BioTechne Grant Foundation).
Author information
Authors and Affiliations
Contributions
G.G.S., F.A., D.T., K.M.F., E.V., S.Y.K. and T.M.H. performed the experiments. G.G.S. designed the experiments, performed the analyses and wrote the manuscript. F.A. designed the nitrosylation experiments, performed the analysis, isolated the adult mouse ventricular myocytes and performed the contractility studies. X.L. isolated the NRVMs. N.J. managed mouse colonies. H.I.M. performed the PV loop experiments and mouse surgeries. Z.V.W. provided the XBP1s transgenic mice and XBP1s adenovirus. P.P.A.M. and J.H. performed the skeletal muscle experiments, D.I.L., V.S.H., K.S. and D.A.K. provided human data. S.L. and T.G.G. contributed to the experimental design and manuscript preparation. J.A.H. conceived the project and contributed to manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
G.G.S., T.G.G. and J.A.H. are co-inventors on a patent application (PCT/US/2017/037019) that was filed in June 2017 (provisional application filed in June 2016). The patent relates to the diet used for modeling HFpEF.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Systemic and cardiac phenotype of mice after 5 or 15 weeks of different dietary regimens.
a, Body weight of mice from different experimental groups after 5 or 15 weeks of diet (n = 10 mice per group for each time point). b, Intraperitoneal glucose-tolerance test (ipGTT) after 5 or 15 weeks of diet (5 weeks, n = 10 mice per group; 15 weeks, n = 5 mice per group). c, Area under the curve of the ipGTT experiment at 5 and 15 weeks (5 weeks, n = 10 mice per group; 15 weeks, n = 5 mice per group). d, e, Systolic blood pressure (SBP) (d) and diastolic blood pressure (DBP) (e) of different experimental groups after 5 or 15 weeks of treatment (n = 10 mice per group for each time point). f, Percentage of LVEF. g, Left ventricular global longitudinal strain. h, Ratio between mitral E wave and A wave. i, Ratio between mitral E wave and E′ wave. j, Ratio between wet and dry lung weight. k, Ratio between heart weight and tibia length. l, Runninng distance during exercise exhaustion test of mice after five weeks of diet. f, h–l, n = 10 mice per group. g, n = 5 mice per group. Data are mean ± s.e.m. a, c–l, One-way ANOVA followed by Sidak’s multiple-comparisons test; numbers above square brackets show significant P values. b, Two-way ANOVA followed by Sidak’s multiple-comparisons test; P values are as follows: 5 weeks 15 min, ***P = 0.0003 chow versus HFD, ***P = 0.0004 chow versus HFD + l-NAME; 30 min ***P = 0.0008 chow versus HFD, **P = 0.006 chow versus HFD + l-NAME; 45 min *P = 0.010 chow versus HFD, ***P = 0.0008 chow versus HFD + l-NAME; 60 min *P = 0.049 chow versus HFD, **P = 0.0096 chow versus HFD + l-NAME; 5 weeks 15 min **P = 0.008 chow versus HFD, ****P < 0.0001 chow versus HFD + l-NAME; 30 min **P = 0.005 chow versus HFD, ****P < 0.0001 chow versus HFD + l-NAME; 45 min **P = 0.009 chow versus HFD, ****P < 0.0001 chow versus HFD + l-NAME; 60 min *P = 0.028 chow versus HFD, **P = 0.0020 chow versus HFD + l-NAME.
Extended Data Fig. 2 Heart morphology and vascular characterization of mice after five weeks of different dietary regimens.
a, Representative images of haematoxylin and eosin (H&E), WGA, Masson’s trichrome (MT) and lectin staining in transversal sections of left ventricle of mice of different experimental groups. Images are representative of four independently performed experiments with similar results. Scale bars, 500 μm (haematoxylin and eosin) and 50 μm (WGA, Masson’s trichrome and lectin). b, WGA quantification of cardiomyocyte cross-sectional area (n = 4 mice per group). c, Percentage of fibrosis area in Masson’s trichrome-stained transversal sections (n = 4 mice per group). d, Myocardial capillary density (n = 4 mice per group). e, Aortic PWV of mice of different experimental groups (n = 5 mice per group). f, Representative pulsed-wave Doppler tracings of coronary flow in mice fed with chow (top) or HFD + l-NAME (bottom) under basal conditions (left; 1.5% isofluorane (Iso)) and after hyperaemic stimulus (right; 3% isofluorane). Images are representative of five independent mice. g, Coronary flow reserve (CFR) quantification (n = 5 mice per group). Data are mean ± s.e.m. b–d, e, g, One-way ANOVA followed by Sidak’s multiple-comparisons test. Numbers above square brackets show significant P values.
Extended Data Fig. 3 Histological and functional analyses of skeletal muscle in mice after five weeks of different dietary regimens.
a, Representative images of haematoxylin and eosin, WGA, metachromatic ATPase (ATPase), Masson’s trichrome and picrosirius red (PSR) staining of soleus and gastrocnemius/plantaris (G/P) muscles from mice fed with chow or HFD + l-NAME. Images are representative of three independently performed experiments with similar results. Scale bars, 50 μm. b, mRNA level of Myhc isoforms (MyHC-1, MyHC-2A or MyHC-2X) of soleus and gastrocnemius/plantaris muscles from mice fed with chow or HFD + l-NAME (n = 5 mice per group). c, Relaxation curve of isolated soleus muscles from mice fed with chow or HFD + l-NAME (n = 5 mice per chow group; n = 6 mice per HFD + l-NAME group). d, e, In vivo forelimb (d) and hindlimb (e) grip force measurements of mice from different experimental groups (n = 8 mice per chow group; n = 4 mice per HFD group; n = 3 mice per l-NAME group; n = 6 mice per HFD + l-NAME group). f, Maximal tetanic stresses in soleus muscles from chow and HFD + l-NAME mice (n = 5 mice per chow group; n = 6 mice per HFD + l-NAME group). Data are mean ± s.e.m. b, c, f, Two-tailed unpaired Student’s t-test (chow versus HFD + l-NAME soleus; chow versus HFD + l-NAME gastrocnemius/plantaris). d, e, One-way ANOVA followed by Sidak’s multiple-comparisons test. Numbers above square brackets show significant P values.
Extended Data Fig. 4 AMVM contractility after 5 weeks of different dietary regimens and functional characterization and analysis of the IRE1α–XBP1 axis in ZSF1-obese 20-week-old rats.
a, Baseline sarcomere length. n = 4 mice and n = 35 cells per chow group; n = 3 mice and n = 30 cells per HFD group; n = 3 mice and n = 30 cells per l-NAME group; n = 3 mice and n = 31 cells per HFD + l-NAME group. b, Time to peak. n = 4 mice and n = 36 cells per chow group; n = 3 mice and n = 30 cells per HFD group; n = 3 mice and n = 30 cells per l-NAME group; n = 3 mice and n = 31 cells per HFD + l-NAME group. c, Maximum return velocity. n = 4 mice and n = 36 cells per chow group; n = 3 mice and n = 29 cells per HFD group; n = 3 mice and n = 30 cells per l-NAME group; n = 3 mice and n = 31 cells per HFD + l-NAME group. d, Change (Δ) in sarcomere length compared to baseline. n = 4 mice and n = 36 cells per chow group; n = 3 mice and n = 30 cells per HFD group; n = 3 mice and n = 30 cells per l-NAME group; n = 3 mice and n = 31 cells per HFD + l-NAME group. e, Maximum departure velocity. n = 4 mice and n = 35 cells per chow group; n = 3 mice and n = 30 cells per HFD group; n = 3 mice and n = 29 cells per l-NAME group; n = 3 mice and n = 28 cells per HFD + l-NAME group. f, Representative tracings of cardiomyocyte contraction–relaxation during pacing. Each trace depicts one cell representative of the average for each experimental group. g–k, Measurements in 20-week old WKY and ZSF1-obese rats. g, Body weight. h, Percentage LVEF. i, Ratio between mitral E wave and A wave. j, Ratio between mitral E wave and E′ wave. k, Ratio between heart weight and tibia length. l, Lung weight to tibia length ratio. g–i, l, n = 5 rats per group; k, n = 5 rats per WKY group and n = 4 rats per ZSF1-obese group. m, Electrophoretic analysis of spliced and unspliced Xbp1 transcripts in left ventricular samples from WKY and ZSF1-obese rats. Tunicamycin-treated NRVMs were used as positive control. n = 3 rats per group. n, Xbp1s mRNA levels in the left ventricles of WKY and ZSF1-obese rats. n = 5 rats per group. o, Immunoblots of p-IRE1α, IRE1α and GAPDH from left ventricular samples of WKY and ZSF1-obese rats. n = 5 rats per group. p, Densitometric analysis of the ratio of p-IRE1α to IRE1α bands. n = 5 rats per group. Data are mean ± s.e.m. a–e, One-way ANOVA followed by Sidak’s multiple-comparisons test. g–l, n, p, Two-tailed unpaired Student’s t-test. Numbers above square brackets show significant P values. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 5 Functional characterization and UPR activation in mice after one week of sTAC.
a, Experimental design. C57BL/6N male mice were exposed to sham (control) or sTAC surgery and were evaluated after 1 week. b, Percentage of LVEF. c, Ratio between mitral E wave and A wave. d, Ratio between mitral E wave and E′ wave. e, Ratio between wet and dry lung weight. f, Ratio between heart weight and tibia length of sham control and sTAC mice. n = 5 mice per group. g, Xbp1s mRNA level in left ventricles of sham and sTAC mice. n = 4 mice in sham group; n = 5 mice in sTAC group. h, Immunoblots of GRP94, GRP78 and GAPDH in left ventricular samples of sham and sTAC mice. Data are representative of three independently performed experiments with similar results. Arrow indicates the regulated band. i, KDEL (Lys-Asp-Glu-Leu) sequence immunofluorescence staining in left ventricular sections of sham and sTAC mice. Hoechst, nuclei. Scale bars, 50 μm. Images are representative of three independently performed experiments with similar results. Data are mean ± s.e.m. b–g, Two-tailed unpaired Student’s t-test. Numbers above square brackets show significant P values. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 6 Functional characterization and analysis of the IRE1α–XBP1 axis in mice after TAC and phenotype of cardiomyocyte-restricted Xbp1s transgenic mice.
a, Experimental design. C57BL/6N mice were exposed to sham (control) or TAC surgery and followed up to five weeks. b–f, Analyses of different experimental groups of mice after sham or before (pre-TAC) and after TAC for one, three or five weeks. b, Ratio between mitral E wave and A wave. c, Ratio between mitral E wave and E′ wave. d, Percentage of LVEF. e, Ratio between heart weight and tibia length. f, Ratio between lung weight and body weight. b–d, n = 10 mice per group for pre-TAC, TAC 1 week and TAC 3 weeks groups; n = 5 mice for the TAC 5 weeks group; e, f, n = 10 mice per group for TAC 1 week and TAC 3 weeks groups; n = 5 mice per group for sham and TAC 5 week groups. g, Xbp1s mRNA levels in the left ventricles of sham-operated mice and mice with TAC for three weeks. n = 5 mice per group. h, Immunoblots of p-IRE1α, IRE1α and GAPDH from left ventricular samples of sham-operated mice and mice with TAC for three weeks. n = 3 mice per group. i, Experimental design. Control (CTR) and Xbp1s transgenic mice (TG) were fed with chow or HFD + l-NAME (green triangle). After five weeks, echocardiographic assessment was performed and doxycycline (Doxy) was removed from the drinking water to induce transgene expression (grey triangle). Two weeks after transgene induction (blue triangle), mice were subjected to functional analysis and tissue collection. j, Percentage of LVEF of different experimental cohorts over time. n = 5 mice per chow control and chow transgenic groups; n = 7 mice per HFD + l-NAME control and HFD + l-NAME transgenic groups. Each mouse was analysed at all three time points. k, Xbp1s mRNA level in left ventricles of control and Xbp1s transgenic mice fed with chow or HFD + l-NAME for seven weeks. n = 3 mice per group. l, Nppa and Nppb left ventricular mRNA levels. m, Ratio between heart weight and tibia length at the end of the study. n = 5 mice per chow control and chow transgenic groups; n = 7 mice per HFD + l-NAME control and HFD + l-NAME transgenic groups. Data are mean ± s.e.m. b–f, One-way ANOVA followed by Sidak’s multiple-comparisons test. g, Two-tailed unpaired Student’s t-test. j–m, Two-way ANOVA followed by Sidak’s multiple-comparisons test. Numbers above square brackets show significant P values. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 7 Myocardial nitrosative stress and inflammatory markers in mice after five weeks of different dietary regimens.
a, Top, cytokine/chemokine antibody array in plasma samples from chow and HFD + l-NAME-treated mice with visual estimation of differently abundant cytokines. Membranes are representative of two independently performed experiments with similar results. Bottom, list of cytokines and chemokines represented by the antibody array membrane. b, Tnf, Il1b and Il6 mRNA levels in left ventricles of mice fed with chow or HFD + l-NAME. n = 5 mice per group. c, iNOS and GAPDH in left ventricular samples of mice fed with chow or HFD + l-NAME. NRVMs infected with mouse AdiNOS (MOI = 100) were used as positive controls for iNOS bands. Arrows indicate the regulated bands. n = 3 mice per group. d, Nos3 (eNOS) mRNA levels in left ventricles from different experimental groups of mice. n = 4 mice per chow group; n = 6 mice per group for HFD, l-NAME and HFD + l-NAME groups. e, Immunoblots of nNOS, eNOS and GAPDH of different experimental groups of mice (n = 3 mice per group). f, Immunoblots of nitrosylated cysteines (Cys-SNO) and GAPDH in left ventricular samples of wild-type and Nos2 knockout (iNOS KO) mice after five weeks of chow or HFD + l-NAME diet. W/B, without blocking. n = 3 mice per group. g, Densitometric analysis of Cys-SNO:GAPDH ratios. n = 3 mice per group. Data are mean ± s.e.m. b, Two-tailed unpaired Student’s t-test. d One-way ANOVA followed by Sidak’s multiple-comparisons test. g, Two-way ANOVA followed by Sidak’s multiple-comparisons test. Numbers above square brackets show significant P values. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 8 iNOS overexpression in cardiomyocytes reduces IRE1α activation and XBP1s levels without affecting cardiomyocyte viability.
a, Immunoblots of iNOS and GAPDH proteins of NRVMs infected with increasing MOIs of AdLacZ or AdiNOS for 24 h. Blots are representative of three independently performed experiments with similar results. b, Nos2 (iNOS) mRNA levels in NRVMs transduced with increasing MOIs of AdLacZ or AdiNOS for 24 h. n = 4 biologically independent experiments. c, Nitrite/nitrate concentration in the medium from NRVMs transduced with increasing MOIs of AdLacZ or AdiNOS for 24 h. n = 4 biologically independent experiments. d, LDH release in NRVMs transduced with increasing MOIs of AdLacZ or AdiNOS for 24 h. n = 3 biologically independent experiments. e, Immunoblots of Cys-SNO, iNOS and GAPDH in NRVMs transduced with AdLacZ or AdiNOS for 24 h (MOI = 100). Blots are representative of three independently performed experiments with similar results. f, Immunoblots of p-IRE1α, IRE1α, iNOS and GAPDH in NRVMs transduced with increasing MOIs of AdLacZ or AdiNOS in the presence or absence of tunicamycin for 24 h. Blots are representative of three independently performed experiments with similar results. g, Xbp1s mRNA level of NRVMs transduced with MOI of 100 of AdLacZ or AdiNOS in the presence or absence of tunicamycin for 24 h. n = 3 biologically independent experiments. Data are mean ± s.e.m. b–d, g, One-way ANOVA followed by Sidak’s multiple-comparisons test. Numbers above square brackets show significant P values. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 9 Phenotype of Nos2 knockout mice after five weeks of different dietary regimens.
a, DNA genotyping of wild-type and Nos2 knockout mice. This signature was used for genotyping. b, Experimental design. Wild-type and Nos2 knockout mice were fed a chow or HFD + l-NAME diet for five weeks. Subsequently, mice were subjected to functional analysis and tissues were collected. c, Percentage of LVEF. d, Body weight.e, Systolic blood pressure. f, Diastolic blood pressure. g, Intraperitoneal glucose-tolerance test of different experimental groups of mice. c, n = 10 mice per group; d–g, n = 5 mice per group. h, Area under the curve of the intraperitoneal glucose-tolerance tests. n = 5 mice per group. i, Ratio between heart weight and tibia length of different experimental groups of mice. n = 5 mice per group. Data are mean ± s.e.m. c–i, Two-way ANOVA followed by Sidak’s multiple-comparisons test. c–f, h, i, Numbers above square brackets show significant P values. g, 15 min ***P = 0.0002 chow wild type versus HFD + l-NAME wild type, ****P < 0.0001 chow wild type versus HFD + l-NAME Nos2 knockout; 30 min ***P = 0.0002 chow wild type versus HFD + l-NAME wild type, ****P < 0.0001 chow wild type versus HFD + l-NAME Nos2 knockout; 45 min ****P < 0.0001 chow wild type versus HFD + l-NAME wild type, ***P = 0.0002 chow wild type versus HFD + l-NAME Nos2 knockout; 60 min ****P < 0.0001 chow wild type versus HFD + l-NAME wild type, ****P < 0.0001 chow wild type versus HFD + l-NAME Nos2 knockout; 120 min **P = 0.010 chow wild type versus HFD + l-NAME wild type, **P = 0.007 chow wild type versus HFD + l-NAME Nos2 knockout. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 10 Functional characterization and myocardial levels of XBP1s in mice treated with an iNOS inhibitor.
a, Experimental design. C57BL/6N mice were fed a chow or HFD + l-NAME diet (brown triangle) for five weeks and subsequently injected intraperitoneally (i.p.) with l-NIL at the dose of 80 mg kg−1 body weight or vehicle twice a day for three days (blue triangles). After that point, mice were subjected to functional analysis and tissues were collected (red triangle). b, Urinary nitrite/nitrate concentration in mice fed a HFD + l-NAME diet treated with vehicle or l-NIL. n = 5 mice per group. c, Systolic blood pressure. d, Diastolic blood pressure. e, Percentage of LVEF. f, Ratio between mitral E wave and A wave. g, Ratio between mitral E wave and E′ wave. h, Running distance during exercise exhaustion test. i, Xbp1s mRNA levels in left ventricles of mice from different experimental groups. b–h, n = 5 mice group; i, n = 3 per group. Data are mean ± s.e.m. b, Two-tailed unpaired Student’s t-test. c–i, Two-way ANOVA followed by Sidak’s multiple-comparisons test. Numbers above square brackets show significant P values.
Supplementary information
Supplementary Figures
This file contains uncropped gel data scans.
Source data
Rights and permissions
About this article
Cite this article
Schiattarella, G.G., Altamirano, F., Tong, D. et al. Nitrosative stress drives heart failure with preserved ejection fraction. Nature 568, 351–356 (2019). https://doi.org/10.1038/s41586-019-1100-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1100-z
This article is cited by
-
Inhibition of ferroptosis reverses heart failure with preserved ejection fraction in mice
Journal of Translational Medicine (2024)
-
Redox proteomics of PANC-1 cells reveals the significance of HIF-1 signaling protein oxidation in pancreatic ductal adenocarcinoma pathogenesis
Journal of Translational Medicine (2024)
-
Linking immune modulation to cardiac fibrosis
Nature Cardiovascular Research (2024)
-
S-nitrosothiol homeostasis maintained by ADH5 facilitates STING-dependent host defense against pathogens
Nature Communications (2024)
-
Targeting HDAC6 to treat heart failure with preserved ejection fraction in mice
Nature Communications (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.