Abstract
The pancreas originates from two epithelial evaginations of the foregut, which consist of multipotent epithelial progenitors that organize into a complex tubular epithelial network. The trunk domain of each epithelial branch consists of bipotent pancreatic progenitors (bi-PPs) that give rise to both duct and endocrine lineages, whereas the tips give rise to acinar cells1. Here we identify the extrinsic and intrinsic signalling mechanisms that coordinate the fate-determining transcriptional events underlying these lineage decisions1,2. Single-cell analysis of pancreatic bipotent pancreatic progenitors derived from human embryonic stem cells reveal that cell confinement is a prerequisite for endocrine specification, whereas spreading drives the progenitors towards a ductal fate. Mechanistic studies identify the interaction of extracellular matrix (ECM) with integrin α5 as the extracellular cue that cell-autonomously, via the F-actin–YAP1–Notch mechanosignalling axis, controls the fate of bipotent pancreatic progenitors. Whereas ECM–integrin α5 signalling promotes differentiation towards the duct lineage, endocrinogenesis is stimulated when this signalling cascade is disrupted. This cascade can be disrupted pharmacologically or genetically to convert bipotent pancreatic progenitors derived from human embryonic stem cells to hormone-producing islet cells. Our findings identify the cell-extrinsic and intrinsic mechanotransduction pathway that acts as gatekeeper in the fate decisions of bipotent pancreatic progenitors in the developing pancreas.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The gene expression datasets generated from sorting E15.5 bipotent and endocrine progenitors (Extended Data Fig. 10e, f) have been deposited in the ArrayExpress database under accession code E-MTAB-6891.
References
Shih, H. P., Wang, A. & Sander, M. Pancreas organogenesis: from lineage determination to morphogenesis. Annu. Rev. Cell Dev. Biol. 29, 81–105 (2013).
Pan, F. C. & Wright, C. Pancreas organogenesis: from bud to plexus to gland. Dev. Dyn. 240, 530–565 (2011).
Ameri, J. et al. Efficient generation of glucose-responsive beta cells from isolated GP2+ human pancreatic progenitors. Cell Reports 19, 36–49 (2017).
Rezania, A. et al. Maturation of human embryonic stem cell-derived pancreatic progenitors into functional islets capable of treating pre-existing diabetes in mice. Diabetes 61, 2016–2029 (2012).
Kilian, K. A., Bugarija, B., Lahn, B. T. & Mrksich, M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc. Natl Acad. Sci. USA 107, 4872–4877 (2010).
Mosqueira, D. et al. Hippo pathway effectors control cardiac progenitor cell fate by acting as dynamic sensors of substrate mechanics and nanostructure. ACS Nano 8, 2033–2047 (2014).
Warmflash, A., Sorre, B., Etoc, F., Siggia, E. D. & Brivanlou, A. H. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods 11, 847–854 (2014).
Dupont, S. et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011).
Cebola, I. et al. TEAD and YAP regulate the enhancer network of human embryonic pancreatic progenitors. Nat. Cell Biol. 17, 615–626 (2015).
George, N. M., Day, C. E., Boerner, B. P., Johnson, R. L. & Sarvetnick, N. E. Hippo signaling regulates pancreas development through inactivation of Yap. Mol. Cell. Biol. 32, 5116–5128 (2012).
Hingorani, S. R. et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4, 437–450 (2003).
Zhang, N. et al. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev. Cell 19, 27–38 (2010).
Kopp, J. L. et al. Sox9+ ductal cells are multipotent progenitors throughout development but do not produce new endocrine cells in the normal or injured adult pancreas. Development 138, 653–665 (2011).
Liu-Chittenden, Y. et al. Genetic and pharmacological disruption of the TEAD–YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 26, 1300–1305 (2012).
Gao, T. et al. Hippo signaling regulates differentiation and maintenance in the exocrine pancreas. Gastroenterology 144, 1543–1553 (2013).
Jansson, L. & Larsson, J. Normal hematopoietic stem cell function in mice with enforced expression of the Hippo signaling effector YAP1. PLoS ONE 7, e32013 (2012).
Holland, A. M. & Hale, M. a, Kagami, H., Hammer, R. E. & MacDonald, R. J. Experimental control of pancreatic development and maintenance. Proc. Natl Acad. Sci. USA 99, 12236–12241 (2002).
Zhao, B. L. L. and K.-L. G. Hippo signaling at a glance. J. Cell Sci. 126, 2135–2140 (2010).
Murtaugh, L. C., Stanger, B. Z., Kwan, K. M. & Melton, D. A. Notch signaling controls multiple steps of pancreatic differentiation. Proc. Natl Acad. Sci. USA 100, 14920–14925 (2003).
Hald, J. et al. Activated Notch1 prevents differentiation of pancreatic acinar cells and attenuate endocrine development. Dev. Biol. 260, 426–437 (2003).
Totaro, A. et al. YAP/TAZ link cell mechanics to Notch signalling to control epidermal stem cell fate. Nat. Commun. 8, 15206 (2017).
Lee, J. C. et al. Regulation of the pancreatic pro-endocrine gene Neurogenin3. Diabetes 50, 928–936 (2001).
Beyer, T. A. et al. Switch enhancers interpret TGF-β and Hippo signaling to control cell fate in human embryonic stem cells. Cell Reports 5, 1611–1624 (2013).
Kim, M., Kim, T., Johnson, R. L. & Lim, D.-S. Transcriptional co-repressor function of the Hippo pathway transducers YAP and TAZ. Cell Reports 11, 270–282 (2015).
Horn, S. et al. Mind bomb 1 is required for pancreatic β-cell formation. Proc. Natl Acad. Sci. USA 109, 7356–7361 (2012).
Halder, G., Dupont, S. & Piccolo, S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat. Rev. Mol. Cell Biol. 13, 591–600 (2012).
Zhao, B. et al. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 26, 54–68 (2012).
Tian, B., Gabelt, B. T., Kaufman, P. L. & Geiger, B. in Encyclopedia of the Eye (ed. Dartt, D. A.) 549–555 (Academic, Cambridge, 2010).
Afrikanova, I. et al. Inhibitors of Src and focal adhesion kinase promote endocrine specification: impact on the derivation of β-cells from human pluripotent stem cells. J. Biol. Chem. 286, 36042–36052 (2011).
Huveneers, S. & Danen, E. H.J. Adhesion signaling—crosstalk between integrins, Src and Rho. J. Cell Sci. 122, 1059–1069 (2009).
Cirulli, V. et al. Expression and function of αVβ3 and αVβ5 integrins in the developing pancreas: roles in the adhesion and migration of putative endocrine progenitor cells. J. Cell Biol. 150, 1445–1459 (2000).
Zhou, Q. et al. A multipotent progenitor domain guides pancreatic organogenesis. Dev. Cell 13, 103–114 (2007).
Seymour, P. A. et al. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc. Natl Acad. Sci. USA 104, 1865–1870 (2007).
Kim, Y. H. et al. Cell cycle-dependent differentiation dynamics balances growth and endocrine differentiation in the pancreas. PLoS Biol. 13, e1002111 (2015).
Klinck, R. et al. A BAC transgenic Hes1-EGFP reporter reveals novel expression domains in mouse embryos. Gene Expr. Patterns 11, 415–426 (2011).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Rezania, A. et al. Enrichment of human embryonic stem cell-derived NKX6.1-expressing pancreatic progenitor cells accelerates the maturation of insulin-secreting cells in vivo. Stem Cells 31, 2432–2442 (2013).
Löf-Öhlin, Z. M. et al. EGFR signaling controls cellular fate and pancreatic organogenesis by regulating apicobasal polarity. Nat. Cell Biol. 19, 1313–1325 (2017).
Rezania, A. et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 32, 1121–1133 (2014).
Kesavan, G. et al. Cdc42/N-WASP signaling links actin dynamics to pancreatic β cell delamination and differentiation. Development 141, 685–696 (2014).
Kesavan, G. et al. Cdc42-mediated tubulogenesis controls cell specification. Cell 139, 791–801 (2009).
Rodríguez-Fraticelli, A. E. et al. Developmental regulation of apical endocytosis controls epithelial patterning in vertebrate tubular organs. Nat. Cell Biol. 17, 241–250 (2015).
Mamidi, A. et al. Signaling crosstalk between TGFβ and Dishevelled/Par1b. Cell Death Differ. 19, 1689–1697 (2012).
Dupont, S. et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFβ signaling, controls Smad4 monoubiquitination. Cell 136, 123–135 (2009).
Funa, N. S. et al. β-catenin regulates primitive streak induction through collaborative interactions with SMAD2/SMAD3 and OCT4. Cell Stem Cell 16, 639–652 (2015).
Stein, C. et al. YAP1 exerts its transcriptional control via TEAD-mediated activation of enhancers. PLoS Genet. 11, e1005465 (2015).
Parreno, J. et al. Expression of type I collagen and tenascin C is regulated by actin polymerization through MRTF in dedifferentiated chondrocytes. FEBS Lett. 588, 3677–3684 (2014).
Schiller, H. B. et al. β1- and αv-class integrins cooperate to regulate myosin II during rigidity sensing of fibronectin-based microenvironments. Nat. Cell Biol. 15, 625–636 (2013).
Acknowledgements
We thank D. Pan for Yap1 floxed mice, L. Jansson and J. Larsson for Tet-O-Yap1 mice, Y. H. Kim and A. Grapin-Botton for Ngn3-RFP mice, Beta Cell Biology Consortium (1 UO1 DK089570-01) for supplying antibodies, J. P. Larsen and S. S. E. Christine for assistance with human pluripotent stem cell expansion and differentiation experiments, G. de la Cruz and P. van Dieken (DanStem FACS core facility), J. Bulkescher (DanStem Imaging core facility), and G. Karemore for assistance with image analysis and statistics. We thank D. Kluver Hansen and A. Stiehm for technical assistance. A.M. is a recipient of a post-doctoral fellowship from Lundbeck Foundation (R151-2013-14359). H.S. and P.S. are recipients of grant HumEn project funded by the European Commission´s Seventh Framework Programme for Research (agreement 602587). P.S. received a grant from Novo Nordisk Foundation (NNF10717), H.S. also received grants from the Lundbeck Foundation (R100-A9422) and the Danish Council for Independent Research (ID: DFF—1331-00310A), Danish Council for Strategic Research. The Novo Nordisk Foundation Center for Stem Cell Biology is supported by a Novo Nordisk Foundation grant number NNF17CC0027852.
Reviewer information
Nature thanks F. Spagnoli and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
A.M., C.P. and H.S. conceived and designed the experiments and wrote the manuscript. P.A.S. performed immunofluorescence staining in Fig. 3c, Extended Data Figs. 3b, 5b, in vivo actin quantifications in Fig. 4b, and Notch mutant analysis in Extended Data Fig. 5e, f. K.H.d.L. performed ChIP experiments in Fig. 3b, Extended Data Fig. 6a and gene expression in Extended Data Fig. 10e, f. A.J. performed staining in Extended Data Fig. 4h. P.S. contributed to materials and data analysis.
Corresponding author
Ethics declarations
Competing interests
A.M. and H.S. are named as inventors in international patent application (WO2016170067A1), which is based on this work. The other authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Validation of hESC-derived pancreatic progenitor differentiation.
a, Schematic of pancreatic progenitor differentiation protocol using the HUES4 hESC PDX1–GFP reporter line. Pancreatic progenitors start to appear from day 10 (D10). b, Expression of endodermal and pancreatic genes from day 0 until day 14. FOXA2 was upregulated early during the definitive endoderm (DE) stage and expression was maintained throughout; this was followed by expression of PDX1 and SOX9, and subsequently NKX6-1. Data from two independent experiments are shown as mean expression ± s.e.m. c, Unsorted pancreatic progenitor culture at day 13 was stained for PDX1 (red), DAPI (blue), GFP (green) and either SOX9, FOXA2, NKX6.1 or YAP1 (grey). Representative images from 4 independent experiments are shown. Scale bar, 200 μm. d, FACS analysis of GFP signal shows transition from GFP− to GFPlow to GFPhigh between days 0 and 17. The red bar represents gating of GFP− (left), GFPlow (middle) and GFPhigh (right). Gating consistency was ensured by the use of flow cytometer calibration beads for EGFP. GFPhigh percentage reached saturation after day 13. Representative FACS plots from 4 independent experiments are shown. e, At day 13, the three populations in d were sorted and analysed by qRT–PCR for gene expression relative to GAPDH. GFPhigh cells were enriched for expression of pancreatic progenitor genes (PDX1, SOX9, NKX6-1, PTF1A and HNF6 (also known as ONECUT1)). There was no change in expression of YAP1 or MKI67. EGFP (n = 5), PDX1 (n = 3), SOX9 (n = 3), FOXA2 (n = 4), NKX6-1 (n = 4), PTF1A (n = 5), HNF6 (n = 3), YAP1 (n = 2), MKI67 (n = 3). Data are mean expression ± s.e.m.
Extended Data Fig. 2 Confinement of single pancreatic progenitor cells is associated with YAP1 downregulation, PDX1 maintenance and endocrine commitment.
a, Time lapse of self-aggregation after adhesion of sorted pancreatic progenitors derived from hESCs. GFP represents PDX1 expression. Representative results from three independent experiments are shown. Scale bar, 100 μm. b, Self-aggregation of sorted human in vitro pancreatic progenitors after 48 h analysed by immunostaining for β-catenin (green) and PDX1 (red), and DAPI staining (blue). Images are representative of three independent experiments. Top left, schematic of self-aggregation experiments to obtain bi-PPs. Top right, illustration of in vivo bi-PPs within trunk epithelium. Bottom left, cluster formed after self-aggregation of sorted bi-PPs. Scale bar, 10 μm. Bottom right, representation of non-spread (PDX1high) and spread (PDX1low) single cells outside the cluster lacking cell–cell contact. Scale bar, 10 μm. c, Cluster from sorted self-aggregated pancreatic progenitor cells stained for pancreatic progenitor markers PDX1 (red), SOX9 or NKX6.1 (grey). β-catenin (green, cell membrane marker), and DAPI (blue). Scale bar, 20 μm. Images are representative of three independent experiments. d, Single cells that are not incorporated into clusters during self-aggregation experiments. Representation of PDX1high confined cells (top row of each pair) and PDX1low spread cells (bottom row of each pair) stained for PDX1 (red), SOX9 (grey), β-catenin (green) and DAPI (blue). Scale bar, 20 μm. Images representative of three independent experiments. e, Sorted single pancreatic progenitor cells adhered on CYTOO chip, stained for F-actin (red) to mark stress fibres and with CellMask (green) to mask cytoplasm, and immunofluorescence for PDX1 (yellow) and SOX9 (grey). Scale bar, 10 μm. Images representative of three independent experiments. f, As in e, but stained for PDX1 (red), NKX6.1 (grey), with CellMask (green), and DAPI (blue). Scale bar, 10 μm. Confined cell (top) and spread cell (bottom). Images representative of three independent experiments. g, Single pancreatic progenitor cells on CYTOO chip 24 h after sorting were analysed by immunostaining and quantified as in Fig. 1a. Nuclear intensity of each pancreatic progenitor marker was quantified and plotted against cell area. Each data point corresponds to an individual cell (PDX1-DISC, n = 309; PDX1-SQUARE, n = 366; NKX6.1, n = 322; HNF6, n = 561; FOXA2, n = 344). Data aggregated from three independent experiments. Spearman’s correlation coefficient (r) is calculated for cell area versus nuclear intensity of staining. PDX1, r = −0.8 (****P ≤ 0.0001); NKX6.1, r = −0.8 (****P ≤ 0.0001); FOXA2, r = −0.1; HNF6, r = −0.2. h, Co-immunofluorescence analysis of 500,000 sorted PDX1–GFPhigh hESC-derived pancreatic progenitor cells, plated and cultured for 72 h on 2-well chamber slides, coated with either fibronectin or laminin, in pancreatic progenitor medium. Cells were immunostained for the ductal markers DBA (white) and SOX9 (red), with DAPI staining (blue). Scale bar, 35 μm. Representative images from three independent experiments are shown. i, Nuclear intensities of YAP1 and NGN3 in individual cells with low spreading (<500 μm2) and high spreading (>500 μm2). <500 μm2, n = 365; >500 μm2, n = 541; 3 independent experiments. Data are shown as mean ± s.e.m.; two-tailed unpaired t-test. j, Expression correlation (r) between PDX1 and YAP1. Top, staining for DAPI (blue), PDX1 (red), YAP1 (grey) and CellMask (green). Scale bar, 20 μm. Nuclear intensity of PDX1 and YAP1 are negatively correlated; r = −0.6 (****P ≤ 0.0001). Each data point corresponds to an individual cell. PDX1/YAP1, n = 460; data aggregated from at least three independent experiments.
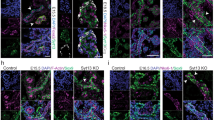
Extended Data Fig. 3 Loss of YAP1 in pancreatic progenitors promotes endocrinogenesis in vivo.
a, qRT–PCR analysis of Yap1 mRNA expression during developmental and early postnatal stages of pancreas organogenesis. Data represented are compared to E11.5 and normalized to HPRT expression. Ordinary one-way ANOVA from three independent embryonic pancreata for each developmental stage; mean ± s.e.m. b, Top, immunofluorescence for YAP1 (red), SOX9 (green) and glucagon (blue) reveals YAP1 expression in SOX9+ pancreatic progenitors but not in glucagon+ endocrine cells at E10.5, supporting western blot analysis in c. Scale bar, 25 µm. Bottom, co-immunofluorescence analysis for SOX9 (white), NGN3 (green), YAP1 (red) and E-cadherin (E-Cad, blue) on sections of E15.5 pancreas. Nuclear YAP1 expression is seen in SOX9+ cells. By contrast, NGN3+ endocrine progenitors (arrows) are either YAP1− or YAP1low with nuclear expression. Scale bar, 5 μm. Images are representative of three embryonic pancreata analysed. c, Western blot of E11.5 pancreatic epithelium (epi.) microdissected from mesenchyme (mes.) and analysed for YAP1 protein expression. Mouse adult islets represent mature endocrine tissue. PDX1 served as control for epithelium and islet expression. YAP1 band intensities were normalized to vinculin. Ordinary one-way ANOVA from three independent experiments; mean ± s.e.m. d, qRT–PCR analysis of TEAD isoforms 1–4 and insulin transcripts from mouse embryonic and early postnatal pancreata. Data represented are compared to E11.5 values and normalized to HPRT expression. Ordinary one-way ANOVA from three independent embryonic pancreata for each developmental stage; *P ≤ 0.1; mean ± s.e.m. e, qRT–PCR and western blot analysis of differentiated, sorted hESCs expressing Ngn3–GFP. Data analysed by ordinary one-way ANOVA; data are mean ± s.e.m. for 3 independent experiments. f, Images of dissected gastrointestinal tracts from P4 control (right), and Pdx1-cre;Yap1fl/fl (left) littermates. Scale bar, 200 μm. Image representative of 10 pups per genotype analysed. Pancreas weight of control and Yap1 KO pups at P4. n = 3 pups analysed. g, Scheme depicting the culture of pancreatic explants ex vivo and representative explants whole-mounted and immunostained for E-cadherin (Ecad, grey) and insulin (red). Images represent 3D reconstructions of confocal images using IMARIS. The ratio of insulin+ area to total epithelial (Ecad+) area was quantified using IMARIS. Explants analysed: wild type, n = 4; Yap1 KO, n = 7; **P = 0.0038. Scale bar, 30 μm. qRT–PCR analysis for Ngn3. Explants analysed: wild type, n = 5; Yap1 KO, n = 5; *P = 0.0364 by two-tailed unpaired t-test. h, Relative gene expression analysis of P4 pancreata for glucagon (Gcg) (wild type, n = 5; Yap1 KO, n = 6; *P = 0.0434), somatostatin (Sst) (wild type, n = 5; Yap1 KO, n = 6; **P = 0.0033), pancreatic polypeptide (Ppy) (wild type, n = 9; Yap1 KO, n = 11; ***P = 0.0008) and ghrelin (Ghrl) (wild type, n = 6; Yap1 KO, n = 7; ***P = 0.0005) by qRT–PCR. Two-tailed unpaired t-test; mean ± s.d. i, qRT–PCR analysis of Sox9-creERT2;Yap1fl/fl explants from g for Yap1 (wild type, n = 5; Yap1 KO, n = 11; ****P = 0.0001), the endocrine genes insulin (wild type, n = 8; Yap1 KO, n = 9; *P = 0.0100), glucagon (wild type, n = 7; Yap1 KO, n = 9; ***P = 0.004), somatostatin (wild type, n = 9; Yap1 KO, n = 8; *P = 0.0023), pancreatic polypeptide (wild type, n = 8; Yap1 KO, n = 9; **P = 0.0013) and Ghrl (wild type, n = 7; Yap1 KO, n = 7; *P = 0.0361) and pancreatic progenitor genes Foxa2 (wild type, n = 7; Yap1 KO, n = 9; **P = 0.0030), Sox9 (wild type, n = 7; Yap1 KO, n = 9; *P = 0.0206), Ptf1a (wild type, n = 8; Yap1 KO, n = 8; **P = 0.0013) and Taz (wild type, n = 9; Yap1 KO, n = 11). Two-tailed unpaired t-test; data are mean ± s.d. j, Wild-type E11.5 pancreata cultured on fibronectin-coated dishes for 5 days, treated during the middle 2 days with 1 μg ml−1 verteporfin (see experimental scheme in g) and analysed for endocrine gene expression. Expression represented relative to HPRT expression levels compared to DMSO-treated explants. n = 4 explants; *P ≤ 0.05; two-tailed unpaired t-test; mean ± s.d.
Extended Data Fig. 4 YAP1 expression levels are critical in governing mouse and human pancreatic progenitor maintenance.
a, Human in vitro pancreatic progenitor cells were transfected with control (Co) or YAP1 siRNA on day 13 and fixed after 72 h, then immunostained for NGN3 (grey), insulin (Ins; red) and DAPI (blue). Scale bar, 42 μm. Images and quantifications of NGN3+ area/DAPI+ area and insulin+ area/DAPI+ area represent four independent experiments. Two-tailed unpaired t-test; mean ± s.e.m. b, hESCs expressing NGN3–GFP, differentiated until S4 stage using the Kieffer protocol and treated with either 1 μg ml−1 verteporfin or DMSO for 2 days and left untreated for another 2 days. Immunostaining for NKX6.1 (grey) and GFP (for NGN3) (green), and stained with DAPI (blue). Scale bar, 70 μm. Images are representative of three independent experiments. NKX6.1+ area/DAPI+ area was quantified using IMARIS. Pooled data from three independent experiments; two-tailed unpaired t-test; mean ± s.d. c, Cells prepared as in b, immunostained for glucagon (grey) and C-peptide (red) with DAPI (blue). Scale bar, 70 μm. Images are representative of three independent experiments. DAPI+ area was quantified using IMARIS. Not significant by two-tailed unpaired t-test; mean ± s.d. d, Wild-type E11.5 mouse pancreatic explants cultured for 5 days ex vivo with 1 μg ml−1 verteporfin or DMSO treatment over days 1–3 and analysed for gene expression after 5 days (Extended Data Fig. 3j). Confocal images of representative explants treated with DMSO (n = 5) or verteporfin (n = 5), immunostained for E-cadherin (Ecad; grey), insulin and glucagon (red), and NGN3 (green) with DAPI (blue). Scale bar, 100 μm. e, E18.5 viscera of control and tet-YAP1S127A;Pdx1tTA/+ (YAP1tg) embryos showing a severe pancreatic agenesis phenotype. YAP1tg P4 pups exhibit severe hyperglycaemia. n = 3; two-tailed unpaired t-test; mean ± s.d. f, Confocal images of E12.5 pancreata from control and YAP1tg littermates immunostained for E-cadherin (Ecad; grey) and YAP1 (green) with DAPI (blue). Scale bar, 33 μm. Images are representative of three pancreata for each genotype. g, qRT–PCR analysis of E12.5 pancreata for known downstream targets of YAP1. Expression is shown relative to HPRT expression. Mean ± s.d.; n = 6 embryos each; two-tailed unpaired t-test (*P ≤ 0.05, **P ≤ 0.01). h, Whole-mount immunostaining for PDX1 (grey) and NKX6.1 (red) of E11.5 control and YAP1tg pancreata. Scale bar, 30 μm. Images are representative of three control and YAP1tg pancreata.
Extended Data Fig. 5 Enforced YAP1 expression promotes acquisition of ductal fate upstream of Notch signalling.
a, qRT–PCR for the pancreatic progenitor markers Pdx1, Sox9, Nkx6-1, Hnf1b, Ngn3 and Cpa1 in E12.5 control and YAP1tg pancreata. Pdx1, Nkx6-1, Ngn3 and Cpa1 are significantly downregulated, but expression of the ductal progenitors Sox9 and Hnf1b is unchanged. Expression is displayed relative to Hprt expression. Mean ± s.e.m.; n = 6 embryos each for control and YAP1tg; two-tailed unpaired t-test. b, Co-immunofluorescence analysis for Muc1 (green), Hnf1β (red) and DBA (grey) with DAPI (blue) on sections of E15.5 pancreata from control or YAP1tg littermates. Insets reveal the boxed regions at higher magnification with the channels separated for clarity below. Ducts, as marked by HNF1β, MUC1 and DBA, are expanded in the YAP1tg (right) compared with control pancreas (left). Scale bar, 100 μm (main panels) or 25 μm (insets). c, qRT–PCR analysis of E15.5 control (Co.) and YAP1tg pancreata reveals upregulation of Sox9 (control, n = 5; YAP1tg, n = 4), Hes1 (control, n = 6; YAP1tg, n = 4) and Notch1 (control, n = 6; YAP1tg, n = 4) transcripts and significant downregulation of endocrine markers Ngn3 (control, n = 6; YAP1tg, n = 4) and Ins1 (control, n = 5; YAP1tg, n = 4) and exocrine genes Cpa1 (control, n = 6; YAP1tg, n = 6) and Amy2a5 (control, n = 5; YAP1tg, n = 4). *P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.0001; two-tailed unpaired t-test; mean ± s.e.m. d, Whole-mount immunostaining of E11.5 + 4 day control and YAP1tg explants cultured and treated with DMSO (control and YAP1tg) or 25 μM γ-secretase inhibitor (YAP1tg). Immunostaining with antibodies against E-cadherin (Ecad; white), insulin (ins) and glucagon (glu) (red) to indicate endocrine differentiation, and CPA1 (green) for acinar differentiation, with DAPI (blue). Scale bar, 60 μm. Note that there is non-specific background with the CPA1 (green) staining outside the epithelium (especially in YAP1tg). Images are representative of n = 5 explants analysed for each condition. e, Confocal optical sections of E10.5 pancreata from Foxa2T2AiCre/+;Rosa26YFP/+ control and Notch signalling mutant (Foxa2T2AiCre/+;Rosa26dnMaml1-eGFP/+) embryos stained for SOX9 (blue), glucagon (grey), GFP (green: shows recombination efficiency in epithelium) and YAP1 (red). Scale bar, 50 μm (main panels), 25 μm (insets). dnMaml1-induced Notch blockade in the pancreatic endoderm did not affect YAP1 expression in SOX9+ progenitors. Images are representative of three pancreata analysed from each genotype. f, YAP1 is normally expressed in the epithelium (GFP+, green) and mesenchyme of E15.5 secondary transition control (Foxa2T2AiCre/+;Rosa26YFP/+) pancreas and its expression in SOX9+ progenitors is unaffected in Notch signalling mutants (Foxa2T2Aicre/+;Rosa26dnMaml1-eGFP/+). SOX9 (grey), YAP1 (red) and GFP (green, shows recombination efficiency in epithelium). Scale bar, 50 μm. Images are representative of three pancreata analysed from each genotype.
Extended Data Fig. 6 YAP1 is an essential mediator of Notch-dependent NGN3 repression
a, ChIP–qPCR of a site 250 bp upstream of the ATG start site of the human NGN3 locus using HES1 antibody. HES1 is specifically enriched at the NGN3 promoter in human in vitro pancreatic progenitor cells, in which NGN3 is repressed. Data represent fold-enrichment over IgG pull-down. Chr14 is used as genomic negative control. Data are mean ± s.e.m. of three independent experiments. b, NGN3 promoter luciferase assay. Left, knockdown of endogenous YAP1 in in vitro pancreatic progenitor cells upregulates NGN3 promoter-driven luciferase activity and the overexpression of exogenous wild-type YAP1, wild-type rat NICD1 or wild-type rat HES1 partially rescues the effects of YAP1 knockdown. n = 3 experiments; *P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.0001; ordinary one-way ANOVA; data are mean ± s.e.m. Middle, western blot analysis of siRNA-treated samples for YAP1 and vinculin loading control. Top right, loss of HES1 results in upregulation of luciferase activity and is partially rescued by overexpression of wild-type YAP1. n = 2 experiments; data are mean ± s.e.m. Bottom right, point mutation of TEAD binding site does not affect luciferase activity, indicating that the binding of YAP1–TEAD repressor complex is not mediated through this site, but the 3× HES1 binding-site mutation significantly upregulates promoter activity. Ordinary one-way ANOVA; n.s, not significant; data are mean ± s.e.m.; n = 6 experiments. c, Co-immunoprecipitation in in vitro pancreatic progenitors. YAP1 forms an endogenous protein complex with TEAD4 and HES1. Likewise, HES1 forms a protein complex with TEAD4 and YAP1, supporting ChIP data on the NGN3 promoter in the pancreatic progenitors. Arrowhead indicates non-specific IgG band. Cartoon depicting a possible YAP1–TEAD–HES1-based transcriptional repressor complex at the NGN3 locus. Representative blots from three independent co-immunoprecipitations performed.
Extended Data Fig. 7 Actin and FAK signalling modulate pancreatic progenitor cell fate via YAP1.
a, Representative western blots for F-actin and G-actin from hESC-derived NGN3+ (representing endocrine precursor cells) and NGN3− (representing pancreatic progenitor cells) sorted at the endocrine precursor stage. *P ≤ 0.05; two-tailed unpaired t-test; data are mean ± s.e.m.; n = 4 independent experiments. b, Scheme depicting timeline of pancreas explant culture and treatments. Immunoblots and quantifications of tubulin, FAK, pFAK Thr397 and YAP1 proteins in explants treated with DMSO, latB or FAK inhibitor after 24 h. YAP1 and pFAK band intensities are normalized to tubulin. Representative images of 3 independent western blot analyses. *P ≤ 0.05, **P ≤ 0.01; two-tailed unpaired t-test; data are mean ± s.e.m. c, Schematic of explant culture timeline. Single-plane optical section confocal images of whole-mount immunostaining for E-cadherin (Ecad, grey) and insulin and glucagon (red) with DAPI (blue). Scale bar, 100 μm. Images are representative of 5 explants analysed for each condition. Gene expression analysis from similar experiments for glucagon and insulin expression. Expression is shown relative to HPRT expression; *P ≤ 0.05, ***P ≤ 0.001; ordinary one-way ANOVA; mean ± s.e.m. Insulin control, n = 4; latB, n = 4; FAK inhibitor, n = 5; glucagon control, n = 3; latB, n = 4; FAK inhibitor, n = 5. d, Western blot analysis of cells represented in (Extended Data Fig. 8b). Cells treated with latB (1 µM) or FAK inhibitor (3 µM) for 24 h. YAP1 band intensity is normalized to tubulin of control cells. *P ≤ 0.05; two-tailed unpaired t-test; data are mean ± s.e.m.; n = 3 independent experiments.
Extended Data Fig. 8 Endocrine differentiation is negatively influenced by FAK and actin signalling, and is associated with increased laminin expression in vivo.
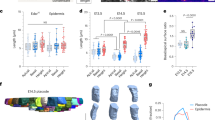
a, Pancreatic progenitors sorted on a dish at single-cell density treated (as in Fig. 4c) with the F-actin inhibitor latB (1 µM) or control (DMSO). Single cells stained for YAP1 and NGN3 and quantified as in Fig. 1a. Each data point representing NGN3+ cells and density corresponds to one experiment. Percentage of NGN3+ cell number represents the number of NGN3+ cells divided by the total number of DAPI+ cells. n.s., non-significant. Two-tailed unpaired t-test; ***P ≤ 0.001; data are mean ± s.e.m. b, Unsorted pancreatic progenitor culture treated with DMSO (control) or latB for 24 h and stained for YAP1 (red), NGN3 (grey) and GFP–PDX1 (green) with DAPI (blue). Scale bar, 20 μm. Representative images of three independent experiments. Image quantification of NGN3 protein is a measure of NGN3+/DAPI+ per cent area. Each data point representing NGN3+ cells and density corresponds to one experiment. **P ≤ 0.01; ***P ≤ 0.001; two-tailed unpaired t-test; data are mean ± s.e.m. Gene expression of NGN3. Each data point corresponds to one experiment. Ordinary one-way ANOVA; mean ± s.e.m. c, Unsorted pancreatic progenitor cultures treated with DMSO (control) or FAK inhibitor for 24 h then stained for YAP1 (red), NGN3 (grey), GFP–PDX1 (green) with DAPI (blue). Scale bar, 20 μm. Images are representative of three independent experiments. d, Quantification of c. NGN3 protein quantification following FAK inhibitor treatment calculated as NGN3+ area/DAPI+ per cent area. Two-tailed unpaired t-test; **P ≤ 0.01; mean ± s.e.m. Each data point corresponds to one experiment. e, Sorted single pancreatic progenitor cells plated on soft substrate (10 kPa) versus glass (≥2,000,000 kPa) coated with fibronectin for 24 h and analysed for percentage NGN3 expression as calculated in a. Percentage of NGN3+ cell number is calculated as the number of NGN3+ cells divided by total number of DAPI+ cells. Data are mean ± s.e.m. of n = 3 experiments; two-tailed unpaired t-test; **P ≤ 0.01. f, Co-immunofluorescence analysis of 30-μm sections from E11.5, E13.5 and E15.5 pancreata for laminin (grey), fibronectin (red) and SOX9 (green) with DAPI (blue). Scale bar, 53 μm. Images are representative of three wild-type pancreata analysed.
Extended Data Fig. 9 Endocrine differentiation is modulated by ECM composition-mediated integrin signalling.
a–e, Pancreatic progenitors sorted at single-cell density on dishes coated with fibronectin, laminin, vitronectin or collagen, fixed after 24 h and stained for PDX1 (fibronectin, n = 436; laminin, n = 252; vitronectin, n = 133; collagen, n = 227 cells), YAP1 (vitronectin, n = 430; collagen, n = 316 cells), NGN3 (vitronectin, n = 300; collagen, n = 205 cells) or YAP1–NGN3 nuclear intensity (vitronectin, n = 300; collagen, n = 216 cells) were quantified as in Fig. 1a. Small blue boxes in e show the YAP1− cell population quantified. Graphs present plots of individual cell data aggregated from three independent experiments. f, Comparison quantification, as shown in Fig. 4c. Each data point representing YAP1+, NGN3+ cells and density corresponds to mean ± s.e.m. of three independent experiments. Percentage of NGN3+ cell number was determined as in Extended Data Fig. 8a. Two-tailed unpaired t-test; ****P ≤ 0.0001, n.s., non-significant; data are mean ± s.e.m. g, Pancreatic progenitor cells sorted as in Fig. 4c and analysed after 24 h culture on vitronectin or collagen. Cell area and nuclear intensities of YAP1+ and NGN3+ cells were measured for cells plated on both ECMs. Two-tailed unpaired t-test; ****P ≤ 0.0001, mean ± s.e.m. of cell data aggregated from three independent experiments. h, i, Co-immunofluorescence and quantification from 30-μm sections from E15.5 wild-type pancreata co-stained for NGN3, fibronectin and laminin (h). NGN3+ cells were segmented using IMARIS surface module. Identification of cells in contact with either laminin only or fibronectin only or both was computed using percentage intensity overlap. The threshold for the intensity overlap was selected manually by examining individual cells. Pie chart showing the percentage of segmented NGN3+ cells in contact with fibronectin or laminin or both. Shown is the mean percentage and confidence interval data of 1,390 NGN3+ cells from 4 independent wild-type E15.5 embryos analysed. i, Pie chart showing the percentage of segmented Sox9+ cells in contact with fibronectin or laminin or both. Shown is the mean percentage and confidence interval data of 2,637 Sox9+ cells from 4 independent wild-type E15.5 embryos analysed. j, Sorted single pancreatic progenitor cells confined (top) or spread (bottom), stained for integrin α5 (ITGa5; red) and NGN3 (grey), with CellMask (green) and DAPI (blue). Scale bar, 10 μm. Representative images from three independent experiments are shown. k, Western blots from sorted hESC-derived cells expressing NGN3–GFP as in Extended Data Fig. 3e. Pancreatic progenitor differentiation performed with the NGN3–GFP reporter line. NGN3high and NGN3low cells (endocrine precursors) express significantly lower levels of integrin α5 (ITGa5) and YAP1 protein than NGN3− cells (pancreatic progenitors). Representative blots from two independent experiments shown. l, Integrin α5 (ITGA5) mRNA expression analysis of NGN3−, NGN3low and NGN3high populations derived from Extended Data Fig. 3e. ITGA5 expression mirrors YAP1 expression during endocrine differentiation (Extended Data Fig. 3e). Expression is presented relative to GAPDH expression. Data are mean ± s.e.m. from two independent differentiation experiments. m, IgG or integrin α5 (ITGa5) antibody staining and flow cytometric analysis of differentiated hESC NGN3–GFP reporter cells. Left, histogram overlay and mean fluorescence intensities (MFI) of IgG-stained (MFI = 100), NGN3–GFP+ endocrine precursor (MFI = 310) and NGN3–GFP− pancreatic progenitor cell (MFI = 704) fractions. Right, staining distribution and MFI of each cell population. NGN3+ endocrine precursors display lower protein expression levels of integrin α5 compared to NGN3− pancreatic progenitors. Data represent three independent experiments. n, mRNA expression analysis of other α integrins (as in l ) that remain unchanged during differentiation of NGN3+ cells from pancreatic progenitors. Expression is shown relative to GAPDH expression; n = 1 experiment.
Extended Data Fig. 10 Pancreatic progenitor cell fate specification is regulated via an ECM–integrin–FAK–actin–YAP1–Notch signalling cascade.
a, Western blot of sorted pancreatic progenitor cells after 24 h adhesion on fibronectin (FN) or laminin (LN). Band intensities were normalized against tubulin. n = 3 independent experiments. pFAK and FAK protein levels are reduced by a similar extent on laminin, indicating that the fraction of pFAK remains constant. Two-tailed unpaired t-tests; *P ≤ 0.05; **P ≤ 0.01, ***P ≤ 0.001; mean ± s.e.m. b, Western blot of sorted pancreatic progenitor cells after 24 h adhesion with control or integrin α5 (ITGa5) inhibition. Band intensities were normalized against GAPDH intensity of control antibody-treated cells. n = 3 independent experiments. pFAK and FAK protein levels are reduced by a similar extent by laminin treatment, indicating that the fraction of pFAK remains constant. Two-tailed unpaired t-tests; *P ≤ 0.05, ***P ≤ 0.001; mean ± s.e.m. c, Unsorted pancreatic progenitor culture transiently transfected with integrin α5 (ITGA5) or control siRNA and analysed after 72 h. Top, NGN3 promoter luciferase assay. Right, gene expression of ITGA5, NGN3, INS and GCG. Each data point corresponds to one experiment. Two-tailed t-tests, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001; mean ± s.e.m. d, Expression of integrin α5 in vivo during secondary transition of mouse embryonic pancreas development. Wild-type E14.5 pancreas stained for integrin α5 (ITGa5; grey) and NGN3 (red) with DAPI (blue). Scale bar, 20 μm. Images are representative of three wild-type pancreata analysed. e, Schematic of sorting mouse E15.5 bi-PPs and endocrine cell populations: Hes1-eGFP;Ngn3-tRFP double reporter mice were dissected and dissociated, stained with DBA and sorted by FACS. Bi-PPs were represented by the DBA+NGN3–tRFP−HES1–EGFP+ population and endocrine precursors were represented by the DBA+ NGN3–tRFP+ HES1–EGFP+ population. All NGN3–tRFP+ cells were also HES1–EGFP+ owing to the long half-life of EGFP. f, Gene expression from microarray analysis of E15.5 FACS-sorted bi-PPs, as in Extended Data Fig. 10e. White bar, DBA+NGN3–tRFP−HES1–EGFP+; grey bar, DBA+NGN3–tRFP+HES1–EGFP+. Shown are all significant differentially expressed (adjusted P values <0.01); n = 4; pools of 5 pancreata. g, Model for bi-PP cell fate specification regulated via ECM–integrin–FAK–actin– YAP1–Notch signalling cascade.
Supplementary information
Supplementary Figures
This file contains the uncropped gel source data scans.
Video 1
: Whole-mount immuno-staining of E12.5 control and Yap Tg pancreata Whole-mount immuno-staining of E12.5 control and Yap Tg pancreata for Sox9 (green) and Muc1 (red) with DAPI (blue); scale bar, 20 μm. Maximum intensity projections of 40x magnification confocal z-stacks are shown for each genotype. Videos are representative of three Control and Yap Tg pancreata analyzed.
Source data
Rights and permissions
About this article
Cite this article
Mamidi, A., Prawiro, C., Seymour, P.A. et al. Mechanosignalling via integrins directs fate decisions of pancreatic progenitors. Nature 564, 114–118 (2018). https://doi.org/10.1038/s41586-018-0762-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0762-2
Keywords
This article is cited by
-
Photo-click hydrogels for 3D in situ differentiation of pancreatic progenitors from induced pluripotent stem cells
Stem Cell Research & Therapy (2023)
-
Mechanically enhanced biogenesis of gut spheroids with instability-driven morphomechanics
Nature Communications (2023)
-
Control of stem cell renewal and fate by YAP and TAZ
Nature Reviews Molecular Cell Biology (2023)
-
The collagen matrix regulates the survival and function of pancreatic islets
Endocrine (2023)
-
Stepwise differentiation of functional pancreatic β cells from human pluripotent stem cells
Cell Regeneration (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.