Abstract
Stimulator of interferon genes (STING) is a receptor in the endoplasmic reticulum that propagates innate immune sensing of cytosolic pathogen-derived and self DNA1. The development of compounds that modulate STING has recently been the focus of intense research for the treatment of cancer and infectious diseases and as vaccine adjuvants2. To our knowledge, current efforts are focused on the development of modified cyclic dinucleotides that mimic the endogenous STING ligand cGAMP; these have progressed into clinical trials in patients with solid accessible tumours amenable to intratumoral delivery3. Here we report the discovery of a small molecule STING agonist that is not a cyclic dinucleotide and is systemically efficacious for treating tumours in mice. We developed a linking strategy to synergize the effect of two symmetry-related amidobenzimidazole (ABZI)-based compounds to create linked ABZIs (diABZIs) with enhanced binding to STING and cellular function. Intravenous administration of a diABZI STING agonist to immunocompetent mice with established syngeneic colon tumours elicited strong anti-tumour activity, with complete and lasting regression of tumours. Our findings represent a milestone in the rapidly growing field of immune-modifying cancer therapies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files). Structure datasets generated during the current study are available in the PDB repository under accession numbers 6DXG and 6DXL. Additional data are available from the corresponding author on reasonable request.
Change history
30 May 2019
Change history: In this Letter, author Ana Puhl was inadvertently omitted; this error has been corrected online.
An amendment to this paper has been published and can be accessed via a link at the top of the paper
References
Corrales, L., McWhirter, S. M., Dubensky, T. W., Jr & Gajewski, T. F. The host STING pathway at the interface of cancer and immunity. J. Clin. Invest. 126, 2404–2411 (2016).
Li, T. & Chen, Z. J. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med. 215, 1287–1299 (2018).
Mullard, A. Can innate immune system targets turn up the heat on ‘cold’ tumours? Nat. Rev. Drug Discov. 17, 3–5 (2018).
Woo, S. R. et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 41, 830–842 (2014).
Klarquist, J. et al. STING-mediated DNA sensing promotes antitumor and autoimmune responses to dying cells. J. Immunol. 193, 6124–6134 (2014).
Diamond, M. S. et al. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J. Exp. Med. 208, 1989–2003 (2011).
Fuertes, M. B. et al. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8alpha+ dendritic cells. J. Exp. Med. 208, 2005–2016 (2011).
Corrales, L. et al. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Reports 11, 1018–1030 (2015).
Conlon, J. et al. Mouse, but not human STING, binds and signals in response to the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid. J. Immunol. 190, 5216–5225 (2013).
Ouyang, S. et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity 36, 1073–1086 (2012).
Zhang, X. et al. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell 51, 226–235 (2013).
Jencks, W. P. On the attribution and additivity of binding energies. Proc. Natl Acad. Sci. USA 78, 4046–4050 (1981).
Ishikawa, H. & Barber, G. N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 (2008).
Ishikawa, H., Ma, Z. & Barber, G. N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792 (2009).
Gao, P. et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153, 1094–1107 (2013).
Diner, E. J. et al. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Reports 3, 1355–1361 (2013).
Gao, P. et al. Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell 154, 748–762 (2013).
Kranzusch, P. J. et al. Ancient origin of cGAS-STING reveals mechanism of universal 2′,3′ cGAMP signaling. Mol. Cell 59, 891–903 (2015).
Shu, C., Yi, G., Watts, T., Kao, C. C. & Li, P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat. Struct. Mol. Biol. 19, 722–724 (2012).
Liu, Y. et al. Activated STING in a vascular and pulmonary syndrome. N. Engl. J. Med. 371, 507–518 (2014).
Melki, I. et al. Disease-associated mutations identify a novel region in human STING necessary for the control of type I interferon signaling. J. Allergy Clin. Immunol. 140, 543–552 (2017).
Jassar, A. S. et al. Activation of tumor-associated macrophages by the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid induces an effective CD8+ T-cell-mediated antitumor immune response in murine models of lung cancer and mesothelioma. Cancer Res. 65, 11752–11761 (2005).
Li, T. et al. Antitumor activity of cGAMP via stimulation of cGAS-cGAMP-STING-IRF3 mediated innate immune response. Sci. Rep. 6, 19049 (2016).
Wang, H. et al. cGAS is essential for the antitumor effect of immune checkpoint blockade. Proc. Natl Acad. Sci. USA 10.1073/pnas.1621363114 (2017).
Acknowledgements
We thank B. Geddes for helpful suggestions and S. Romeril for discussions and comments.
Reviewer information
Nature thanks B. Stockwell and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
J.M.R. conceived the dimer concept and designed compound 2, and conceived the concept for compound 3 and synthetic chemistry for compound 4. G.S.P. identified compound 1. J.M.R., G.S.P. and J.Y. were co-leaders and oversaw the research program. J.M.R., G.S.P and J.Y. wrote the manuscript with assistance from all other authors. N.C. performed HDX studies and determined X-ray structures with assistance from L.W. and A.C.P. R.S. synthesized compounds 2 and 4. S.-Y.Z., M.A., and C.B.H. conducted the in vivo efficacy study in CT-26 tumour-bearing mice. J.-L.T. conducted in vivo pharmacodynamics studies in wild-type and Sting−/− mice. P.M. performed in vitro functional experiments in PBMCs. S.L., H.C.E., M.M., M.B., and G.B. designed, performed and analysed chemoproteomics experiments. M.K. and J.L.S. developed and assisted with the high-throughput screening assay. J.C. conducted PBMC assays from different haplotype donors. J.M., J.R., A.M., L.L., T.L.G., A.K.C., G.Y., and Y. Li contributed to design, optimization of synthetic route and preparation of compounds. N.N. carried out structure-based design analysis. A.I.W., V.K., and P.M. characterized agonist activity. K.N. purified STING protein. J.G. conducted thermal shift experiments. K.B. and M.A.R. designed and supervised pharmacokinetic studies. K.P.F. was co-leader during program initiation. P.J.G. supervised biology and provided advice. Y. Lian, K.J.D., and J.A. contributed to compound selection. R.W.M. contributed to chemistry strategy and provided advice. J.K. contributed to optimization of synthetic route and preparation of compounds. J.S., A.H. and J.B. jointly supervised the program.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
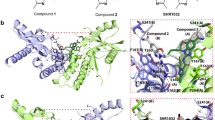
Extended Data Fig. 1 Co-crystal structures and superimposition of compounds 1 and 2.
a, Superposition of compound 1 (PDB code: 6DXG) and the diABZI compound 2 (PDB code: 6DXL) bound to human STING (aa 149–379). b, Intermolecular contacts in the complex of compounds 1 and 2 bound to human STING (aa 149–379). Magenta, compound 1; green, compound 2. Corresponding subunits of STING shown in same colour for compounds 1 and 2. c, Electron density (1.0σ) of compound 1. d, Electron density of (0.5σ) of compound 2.
Extended Data Fig. 2 Selectivity of compound 2 determined by affinity enrichment chemoproteomics.
To identify any potential off-target liabilities early on, an affinity enrichment-based chemoproteomics strategy was applied to compound 2. Compound 5, an active analogue containing a primary amine functionality, was covalently immobilized on sepharose beads and was used to affinity-capture potential target proteins from a THP1 cell lysate. Pull-down experiments were performed in the absence of free compound 2 to delineate target proteins from background or in the presence of compound 2 over a range of concentrations. All proteins captured by the beads under the different conditions were eluted and subsequently quantified by isotope tagging of tryptic peptides followed by LC–MS/MS analysis to establish a competition-binding curve and determine a half-maximal inhibition (IC50) value. The IC50 values obtained in these experiments represent a measure of target affinity, but are also affected by the affinity of the target for the bead-immobilized ligand. The latter effect can be deduced by determining the depletion of the target proteins by the beads, such that apparent dissociation constants (\({K}_{{\rm{d}}}^{{\rm{app}}}\)) can be determined, which are largely independent from the bead ligand (see Supplementary Methods for details). Notably, only two proteins were captured and competed in a dose-dependent manner within a 1,000-fold window, namely STING and orosomucoid1 (ORM1, alpha-1-acid glycoprotein 1 precursor). The mean \({K}_{{\rm{d}}}^{{\rm{app}}}\) value for STING was determined as 1.6 nM, demonstrating high potency of compound 2 on the target protein not only in an artificial biochemical assay system using truncated protein but also against the full-length endogenous human protein. The mean \({K}_{{\rm{d}}}^{{\rm{app}}}\)value of the only identified off-target protein, ORM1, was determined as 79 nM giving a comfortable selectivity window of approximately 40-fold. ORM1 is an acute phase reactant, an abundant plasma protein with known drug binding properties, and is known to be expressed in monocytes.
Extended Data Fig. 3 Superimposition of co-crystal structures of cGAMP and compound 2.
a, Superimposition of bound conformations of cGAMP (yellow) and diABZI compound 2 (green) bound to human STING (aa 149–379). b, Superimposition of bound structures of cGAMP and diABZI compound 2.
Extended Data Fig. 4 Bound conformations of cGAMP and compound 2.
a, Conformations of cGAMP bound to human STING (aa 149–379). b, diABZI compound 2 bound to human STING (aa 149–379).
Extended Data Fig. 5 Anti-CD8 depletion antibody validation by flow cytometry.
a, Schematic of CD8T cell depletion scheme with timings consistent with efficacy studies. b, c, Flow cytometry quantification of CD4 and CD8 T cells from vehicle-treated (b) or anti-CD8 antibody (BioXcell: clone 2.43)-treated (c) BALB/c mice. Blood taken before dosing and after the third dose and spleen samples validate effective depletion of CD8+ T cells. Similar results observed 72 h after dose 1 and dose2. d, Flow cytometry gating strategy. Flow cytometry staining and gating blood samples were collected via tail snip for pre-dose bleeds and via cardiac puncture under isoflurane following the third dose. An equal volume of blood was added to flow staining buffer (PBS + 0.5% BSA), and samples were incubated in mouse Fc blocker. Spleen samples were processed to cell suspension, resuspended in flow staining buffer, and incubated with mouse Fc blocker. Samples were stained with live/dead aqua dye, CD45–PE, CD3–V421, and CD8–APC. Gating strategy reports the percentage positive population of live cells → CD45+ → CD3+. All samples were run on BD Canto II and analysed with FACSDiva software.
Supplementary information
Supplementary Information
This file contains the Supplementary Methods section and the Supplementary information as a combined single document
Rights and permissions
About this article
Cite this article
Ramanjulu, J.M., Pesiridis, G.S., Yang, J. et al. Design of amidobenzimidazole STING receptor agonists with systemic activity. Nature 564, 439–443 (2018). https://doi.org/10.1038/s41586-018-0705-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0705-y
Keywords
This article is cited by
-
Nanomaterial-encapsulated STING agonists for immune modulation in cancer therapy
Biomarker Research (2024)
-
cGAS-STING, inflammasomes and pyroptosis: an overview of crosstalk mechanism of activation and regulation
Cell Communication and Signaling (2024)
-
Exploring the next generation of antibody–drug conjugates
Nature Reviews Clinical Oncology (2024)
-
Activating STING/TBK1 suppresses tumor growth via degrading HPV16/18 E7 oncoproteins in cervical cancer
Cell Death & Differentiation (2024)
-
A conserved ion channel function of STING mediates noncanonical autophagy and cell death
EMBO Reports (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.