Abstract
Somatic mutations in the isocitrate dehydrogenase 2 gene (IDH2) contribute to the pathogenesis of acute myeloid leukaemia (AML) through the production of the oncometabolite 2-hydroxyglutarate (2HG)1,2,3,4,5,6,7,8. Enasidenib (AG-221) is an allosteric inhibitor that binds to the IDH2 dimer interface and blocks the production of 2HG by IDH2 mutants9,10. In a phase I/II clinical trial, enasidenib inhibited the production of 2HG and induced clinical responses in relapsed or refractory IDH2-mutant AML11. Here we describe two patients with IDH2-mutant AML who had a clinical response to enasidenib followed by clinical resistance, disease progression, and a recurrent increase in circulating levels of 2HG. We show that therapeutic resistance is associated with the emergence of second-site IDH2 mutations in trans, such that the resistance mutations occurred in the IDH2 allele without the neomorphic R140Q mutation. The in trans mutations occurred at glutamine 316 (Q316E) and isoleucine 319 (I319M), which are at the interface where enasidenib binds to the IDH2 dimer. The expression of either of these mutant disease alleles alone did not induce the production of 2HG; however, the expression of the Q316E or I319M mutation together with the R140Q mutation in trans allowed 2HG production that was resistant to inhibition by enasidenib. Biochemical studies predicted that resistance to allosteric IDH inhibitors could also occur via IDH dimer-interface mutations in cis, which was confirmed in a patient with acquired resistance to the IDH1 inhibitor ivosidenib (AG-120). Our observations uncover a mechanism of acquired resistance to a targeted therapy and underscore the importance of 2HG production in the pathogenesis of IDH-mutant malignancies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Losman, J. A. & Kaelin, W. G. Jr. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 27, 836–852 (2013).
Ley, T. J. et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456, 66–72 (2008).
Ward, P. S. et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17, 225–234 (2010).
Gross, S. et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J. Exp. Med. 207, 339–344 (2010).
Losman, J. A. et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 339, 1621–1625 (2013).
Figueroa, M. E. et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18, 553–567 (2010).
Kats, L. M. et al. Proto-oncogenic role of mutant IDH2 in leukemia initiation and maintenance. Cell Stem Cell 14, 329–341 (2014).
Chen, C. et al. Cancer-associated IDH2 mutants drive an acute myeloid leukemia that is susceptible to Brd4 inhibition. Genes Dev. 27, 1974–1985 (2013).
Yen, K. et al. AG-221, a first-in-class therapy targeting acute myeloid leukemia harboring oncogenic IDH2 mutations. Cancer Discov. 7, 478–493 (2017).
Wang, F. et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science 340, 622–626 (2013).
Stein, E. M. et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood 130, 722–731 (2017).
Amatangelo, M. D. et al. Enasidenib induces acute myeloid leukemia cell differentiation to promote clinical response. Blood 130, 732–741 (2017).
Gorre, M. E. et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science 293, 876–880 (2001).
Kobayashi, S. et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 352, 786–792 (2005).
Pao, W. et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2, e73 (2005).
Choi, Y. L. et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N. Engl. J. Med. 363, 1734–1739 (2010).
Shih, A. H. et al. Combination targeted therapy to disrupt aberrant oncogenic signaling and reverse epigenetic dysfunction in IDH2- and TET2-mutant acute myeloid leukemia. Cancer Discov. 7, 494–505 (2017).
Niederst, M. J. et al. The allelic context of the C797S mutation acquired upon treatment with third-generation EGFR inhibitors impacts sensitivity to subsequent treatment strategies. Clin. Cancer Res. 21, 3924–3933 (2015).
Zehir, A. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23, 703–713 (2017).
He, J. et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood 127, 3004–3014 (2016).
Yang, S., Shi, H., Chu, X., Zhou, X. & Sun, P. A rapid and efficient polyethylenimine-based transfection method to prepare lentiviral or retroviral vectors: useful for making iPS cells and transduction of primary cells. Biotechnol. Lett. 38, 1631–1641 (2016).
Dang, L. et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744 (2009).
Schrödinger. Schrödinger Release 2017-2: Maestro (Schrödinger, LLC, New York, 2017).
Jacobson, M. P., Friesner, R. A., Xiang, Z. & Honig, B. On the role of the crystal environment in determining protein side-chain conformations. J. Mol. Biol. 320, 597–608 (2002).
Jacobson, M. P. et al. A hierarchical approach to all-atom protein loop prediction. Proteins 55, 351–367 (2004).
Acknowledgements
We thank members of the Levine and Thompson laboratories for discussions. We thank W.K. Chatila and N. Schultz for assistance with data deposition. A.M.I. is supported by the NIH/NCI (K08 CA201483), Leukemia & Lymphoma Society (3356-16), Burroughs Wellcome Fund (1015584), Susan & Peter Solomon Divisional Genomics Program, Steven A. Greenberg Fund, and Cycle for Survival. A.H.S. is supported by the NIH/NCI (K08 CA181507) and Leukemia & Lymphoma Society. The work was also supported, in part, by the Conquer Cancer Foundation of ASCO (A.M.I., A.H.S. and J.T.), the Leukemia & Lymphoma Society Specialized Center of Research Program (7011-16; A.M.I. and C.B.T.), a Translational and Integrative Medicine Research Fund (TIMRF) grant (A.H.S. and E.M.S.), the American Association for Cancer Research (J.T.), the American Society of Hematology/Robert Woods Johnson Foundation (J.T.), and grants from the NIH, including R01 CA168802-02 (C.B.T.), R35 CA197594-01A1 (R.L.L.), U54 OD020355 (R.L.L.), and the Memorial Sloan Kettering Cancer Center Support Grant (NIH P30 CA008748) including a supplement to R.L.L., C.B.T. and A.H.S. We acknowledge the use of the Integrated Genomics Operation Core, funded by the Memorial Sloan Kettering Cancer Center Support Grant (NIH P30 CA008748), Cycle for Survival, and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology.
Reviewer information
Nature thanks J. Cortes and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
A.M.I., A.H.S., C.B.T., R.L.L. and E.M.S. conceived the project, designed the experiments, analysed the data, and wrote the manuscript. A.M.I., A.H.S., B.W. and A.N. performed the experiments with technical assistance from F.M.C., N.T., V.D., H.L. and J.R.C. M.P., C.F., J.T. and M.S.T. assisted with management of clinical data and specimens. M.E.A. and M.R. assisted with pathological assessment of biospecimens. A.S.R., S.K.A., G.A.P. and J.D.C. performed the structural modelling. N.F. and E.P. performed the mutational analysis. B.W., S.C., Z.D.K. and S.A.B. assisted with identification and analysis of the IDH1-mutant leukaemia. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
C.B.T. is a founder of Agios Pharmaceuticals and a member of its scientific advisory board. He also serves on the board of directors of Merck and Charles River Laboratories. R.L.L. is on the Supervisory Board of Qiagen. J.D.C. is a member of the scientific advisory board of Schrödinger. B.W., S.C., Z.D.K. and S.A.B. are employees of Agios Pharmaceuticals, Inc.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Acquired clinical resistance to the mutant IDH2 inhibitor enasidenib (AG-221).
a, b, Haematoxylin and eosin staining of bone marrow cells aspirated from patient A (a) and patient B (b) at indicated points in relation to treatment with AG-221. Remission images demonstrate decreased leukaemic blasts and increased myeloid differentiation that are reversed at the time of relapse. Images show 100× magnification. Images are representative fields of a single bone marrow aspiration performed at each time point.
Extended Data Fig. 2 Structures illustrating potential interactions between IDH2 second-site mutations and enasidenib.
a–h, Detailed view of the interactions between wild-type IDH2 Q316 (a, e) and Q316’ (c, g) or mutant IDH2(Q316E) (b, f) and IDH2(Q316E’) (d, h) with AG-221 in the predicted dominant conformation (a–d) or a minor conformation (e–h). Hydrogen bonds are depicted in light green. Note the disrupted hydrogen bond (depicted as orange bar) in d resulting from the Q316E mutation in the IDH2’ subunit. i–p, Detailed view of the interactions between wild-type IDH2 I319 (i, m) and I319’ (k, o) or mutant IDH2(I319M) (j, n) and IDH2(I319M’) (l, p) with AG-221 in the predicted dominant conformation (i–l) or a predicted minor conformation (m–p). The solvent-excluded surface of AG-221 is shown transparently in grey. The van der Waals radius of the Cδ1 and Cγ2 atoms of I319/I319’ or the Sδ and Cε atoms of I319M/I319M’ are depicted as spheres. Unfavourable steric interactions between AG-221 and these atoms are depicted in red. Throughout the figure, the IDH2 subunit is depicted in blue-grey, the IDH2’ subunit in purple, and AG-221 in teal. Non-polar hydrogen atoms are not shown. White, red, blue and yellow portions of stick structures indicate hydrogen, oxygen, nitrogen and sulfur atoms, respectively. All models were based on the AG-221–IDH2 structure (PDB code 5I96)9 (see Methods).
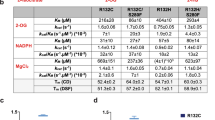
Extended Data Fig. 3 Expression and activity of in trans IDH2 second-site mutations in haematopoietic cells.
a, Allele-specific quantitative PCR (qPCR) showing similar expression of constructs in Ba/F3 cells co-transduced with IDH2R140Q (RQ) plus IDH2WT (WT), IDH2Q316E (QE), or IDH2I319M (IM) in trans. Control from IDH2WT human cell line (293T). Data are mean ± s.e.m. for triplicate reactions. b, Intracellular 2HG levels in Ba/F3 cells co-expressing RQ plus WT, QE or IM in trans and treated with vehicle or increasing doses of AG-221 (1 nM, 10 nM, 100 nM, 1 μM or 10 μM). Data are mean ± s.e.m. for triplicate cultures. c, Western blot showing IDH2 protein levels in primary HSPCs from Idh2R140Q/Flt3ITD mice transduced with WT, QE or IM and untransduced control cells for comparison. GAPDH serves as a loading control. The same membrane was stripped and reprobed for western blots. d, Intracellular 2HG levels in primary HSPCs from Idh2R140Q/Flt3ITD mice transduced with WT, QE or IM and collected from the first passage of methylcellulose cultures containing AG-221 at 50 nM. Data for are mean ± s.e.m. for triplicate cultures. e, Flow cytometry gating strategy for Fig. 3i. SSC-A, side scatter area; FSC-A, forward scatter area. DAPI is a viability dye. mCherry identifies retrovirally transduced cells. Results are representative of ≥2 (a–d) or 1 (e) independent experiments. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 4 Purification and activity of IDH2(R140Q) dimers with wild-type IDH2 or mutants Q316E or I319M in trans.
a, Schematic of experimental approach: 293T cells were co-transfected with HA-tagged IDH2(R140Q) plus Flag-tagged wild-type, Q316E or I319M. After 2 days, cells were lysed and enzyme complexes were purified by HA-immunoprecipitation. Reactions were performed with purified enzyme, NADPH, αKG and varying doses of AG-221 as detailed in Fig. 3. b–e, Purity and dimerization of HA-precipitated enzymes were assessed by denatured SDS–PAGE with Coomassie staining (b), denatured SDS–PAGE with western blotting (c), native PAGE with Coomassie staining (d), or native PAGE with western blotting for the indicated proteins (e). Separate membranes were used for western blots. f, In vitro enzyme assays measuring rate of NADPH consumption of IDH2 dimers purified as in b–e. Reactions contained purified enzyme (10 μg ml−1), NADPH (0.3 mM), αKG (5 mM) and AG-221 at indicated concentrations. Data are mean ± 95% confidence intervals for triplicate reactions. Results are representative of ≥3 (b–d, f) or 2 (e) independent experiments. For gel source data, see Supplementary Fig. 2.
Extended Data Fig. 5 Second-site IDH2 mutations in cis can confer resistance to enasidenib.
a, Western blot showing IDH2 expression in Ba/F3 cells transduced with the indicated constructs. Vinculin serves as a loading control. The same membrane was probed for both IDH2 and vinculin. These are the same cells as in Fig. 4a. b–g, Purification and enzymatic activity of IDH2 WT–R140Q dimers with or without in cis second-site mutations. b, Schematic of experimental approach: 293T cells were co-transfected with HA-tagged wild-type IDH2 plus Flag-tagged IDH2(R140Q), in cis double-mutant IDH2 R140Q/Q316E (RQ/QE) or in cis double-mutant IDH2 R140Q/I319M (RQ/IM). After 2 days, cells were lysed and IDH2 enzyme complexes were purified by HA-immunoprecipitation. c–e, Purity and dimerization of HA-precipitated enzymes were assessed by denatured SDS–PAGE with Coomassie staining (c), denatured SDS–PAGE with western blotting with the indicated antibodies (d), or native PAGE with Coomassie staining (e). Separate membranes were used for Western blots. f, g, In vitro enzyme assays measuring relative activity (f) and rate of NADPH consumption (g) by HA-precipitated IDH2 dimers. Reactions contained purified enzyme (7.5 μg ml−1), NADPH (0.3 mM), αKG (5 mM), and vehicle or increasing doses of AG-221 (0.1, 0.3, 1, 3, 10 or 30 μM). Data are mean ± 95% confidence interval for triplicate reactions (duplicate reactions for WT–RQ/QE AG-221 3 μM and 30 μM). Results are representative of ≥3 independent experiments. For gel source data, see Supplementary Fig. 3.
Supplementary Information
Supplementary Figures 1-3
Supplementary Figure 1 contains full blots for Extended Data Figure 3c, Supplementary Figure 2 contains full blots for Extended Data Figure 4c and 4e and Supplementary Figure 3 contains full blots for Extended Data Figure 5a and 5d.
Supplementary Tables 1-5
Supplementary Table 1 contains RainDance microdroplet-based PCR sequencing results for IDH2 in Patient A (Q316E mutation on enasidenib). Supplementary Table 2 contains RainDance microdroplet-based PCR sequencing results for IDH2 in Patient B (I319M mutation on enasidenib). Supplementary Table 3 contains FoundationOne Heme hybrid-capture sequencing results for IDH1 in Patient X (S280F mutation on ivosidenib). Supplementary Table 4 contains RainDance microdroplet-based PCR sequencing results for IDH2 in patients treated with enasidenib, including those with primary or acquired resistance. Supplementary Table 5 contains MSK-IMPACT hybrid-capture sequencing results for IDH2 (R140, Q316, I319) in patients treated with enasidenib, including those with primary or acquired resistance.
Rights and permissions
About this article
Cite this article
Intlekofer, A.M., Shih, A.H., Wang, B. et al. Acquired resistance to IDH inhibition through trans or cis dimer-interface mutations. Nature 559, 125–129 (2018). https://doi.org/10.1038/s41586-018-0251-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0251-7
This article is cited by
-
IDH2/R140Q mutation confers cytokine-independent proliferation of TF-1 cells by activating constitutive STAT3/5 phosphorylation
Cell Communication and Signaling (2024)
-
Designing patient-oriented combination therapies for acute myeloid leukemia based on efficacy/toxicity integration and bipartite network modeling
Oncogenesis (2024)
-
Therapie der rezidivierten/refraktären akuten myeloischen Leukämie
Im Fokus Onkologie (2023)
-
Update on Small Molecule Targeted Therapies for Acute Myeloid Leukemia
Current Treatment Options in Oncology (2023)
-
Exploration of natural product database for the identification of potent inhibitor against IDH2 mutational variants for glioma therapy
Journal of Molecular Modeling (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.