Abstract
The mechanistic target of rapamycin complex 1 (mTORC1) is a master regulator of cell growth that responds to a diverse set of environmental cues, including amino acids1,2. Deregulation of mTORC1 has been linked with metabolic diseases, cancer and ageing2,3,4. In response to amino acids, mTORC1 is recruited by the Rag GTPases to the lysosome, its site of activation5,6. The GATOR1 complex, consisting of DEPDC5, NPRL3 and NPRL2, displays GAP activity to inactivate Rag GTPases under amino-acid-deficient conditions7. However, it is unclear how the inhibitory function of GATOR1 is released upon amino acid stimulation. Here we find that in response to amino acids, the CUL3–KLHL22 E3 ubiquitin ligase promotes K48-linked polyubiquitination and degradation of DEPDC5, an essential subunit of GATOR1. KLHL22 plays a conserved role to mediate the activation of mTORC1 and downstream events in mammals and nematodes. Depletion of MEL-26, the Caenorhabditis elegans orthologue of KLHL22, extends worm lifespan. Moreover, KLHL22 levels are elevated in tumours of breast cancer patients, whereas DEPDC5 levels are correspondingly reduced. Depletion of KLHL22 in breast cancer cells suppresses tumour growth in nude mice. Therefore, pharmacological interventions targeting KLHL22 may have therapeutic potential for the treatment of breast cancer and age-related diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Dibble, C. C. & Manning, B. D. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat. Cell Biol. 15, 555–564 (2013).
Laplante, M. & Sabatini, D. M. mTOR signaling in growth control and disease. Cell 149, 274–293 (2012).
Yuan, H. X., Xiong, Y. & Guan, K. L. Nutrient sensing, metabolism, and cell growth control. Mol. Cell 49, 379–387 (2013).
Zoncu, R., Efeyan, A. & Sabatini, D. M. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 (2011).
Kim, E., Goraksha-Hicks, P., Li, L., Neufeld, T. P. & Guan, K. L. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 10, 935–945 (2008).
Sancak, Y. et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 (2008).
Bar-Peled, L. et al. A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340, 1100–1106 (2013).
Sabatini, D. M. mTOR and cancer: insights into a complex relationship. Nat. Rev. Cancer 6, 729–734 (2006).
Guertin, D. A. & Sabatini, D. M. Defining the role of mTOR in cancer. Cancer Cell 12, 9–22 (2007).
Wolfson, R. L. et al. KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature 543, 438–442 (2017).
Mertins, P. et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534, 55–62 (2016).
Petroski, M. D. & Deshaies, R. J. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 (2005).
Dhanoa, B. S., Cogliati, T., Satish, A. G., Bruford, E. A. & Friedman, J. S. Update on the Kelch-like (KLHL) gene family. Hum. Genomics 7, 13 (2013).
Pintard, L., Willems, A. & Peter, M. Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J. 23, 1681–1687 (2004).
Beck, J. et al. Ubiquitylation-dependent localization of PLK1 in mitosis. Nat. Cell Biol. 15, 430–439 (2013).
Radivojac, P. et al. Identification, analysis, and prediction of protein ubiquitination sites. Proteins 78, 365–380 (2010).
Wagner, S. A. et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10, M111–013284 (2011).
Peng, M., Yin, N. & Li, M. O. SZT2 dictates GATOR control of mTORC1 signalling. Nature 543, 433–437 (2017).
Jewell, J. L. et al. Metabolism. Differential regulation of mTORC1 by leucine and glutamine. Science 347, 194–198 (2015).
Hosokawa, N. et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 20, 1981–1991 (2009).
Fingar, D. C., Salama, S., Tsou, C., Harlow, E. & Blenis, J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 16, 1472–1487 (2002).
Lapierre, L. R. et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat. Commun. 4, 2267 (2013).
Settembre, C. et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31, 1095–1108 (2012).
Kaeberlein, M. et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310, 1193–1196 (2005).
Vellai, T. et al. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature 426, 620 (2003).
Kapahi, P. et al. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 14, 885–890 (2004).
Harrison, D. E. et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460, 392–395 (2009).
Lerman, M. I. & Minna, J. D. The 630-kb lung cancer homozygous deletion region on human chromosome 3p21.3: identification and evaluation of the resident candidate tumor suppressor genes. The International Lung Cancer Chromosome 3p21.3 Tumor Suppressor Gene Consortium. Cancer Res. 60, 6116–6133 (2000).
Li, J. et al. Functional characterization of the candidate tumor suppressor gene NPRL2/G21 located in 3p21.3C. Cancer Res. 64, 6438–6443 (2004).
Seng, T. J. et al. Complex chromosome 22 rearrangements in astrocytic tumors identified using microsatellite and chromosome 22 tile path array analysis. Genes Chromosom. Cancer 43, 181–193 (2005).
Acknowledgements
We thank S. Perrett and S. Song for proofreading our manuscript. We thank S.-C. Lin, G. Ouyang, Q. Wu and Y. Wang for providing us with reagents. Several C. elegans strains were provided by CGC, which is supported by the NIH-Officer of Research Infrastructure Programs. This work is supported by the Ministry of Science and Technology of China (National Key Research and Development Program of China grant 2017YFA0504000 and 973 grant 2013CB910104), the National Natural Science Foundation of China (31422033), Peking-Tsinghua Center for Life Sciences, and an HHMI International Research Scholar Award (HHMI#55008739) to Y.L.
Reviewer information
Nature thanks F. Basserman and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
J.C., assisted by Y.O., performed most of the experiments and analysed the data. Y.Y. generated the mel-26; HLH-30::GFP strain. W.L. generated DEPDC5 and KLHL22 point mutation constructs. Y.Xu. and Y.Xi. provided breast cancer samples. J.C. and Y.L. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
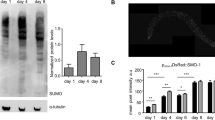
Extended Data Fig. 1 DEPDC5 undergoes ubiquitin-mediated degradation in response to amino acids.
a, Protein levels of endogenous DEPDC5 are regulated in response to the availability of amino acids. Basal, standard culture condition. b, Amino acids do not affect mRNA levels of DEPDC5. Relative mRNA levels of DEPDC5 to GAPDH were quantified by qPCR. n = 3 biologically independent experiments. ns, no significant difference; two-sided Student’s t-test. c, d, DEPDC5 proteins are unstable under normal conditions. CHX, cycloheximide. e, Proteasome inhibitor MG132 increases the protein levels of DEPDC5. f, The ubiquitination of DEPDC5 is regulated in an amino-acid-dependent manner. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 2 CUL3–KLHL22 mediates the ubiquitination of DEPDC5.
a, KLHL22, but not other E3 ligases, promotes the degradation of DEPDC5. b, DEPDC5 interacts with CUL3 and RBX1. Cells stably expressing HA–DEPDC5 were subjected to anti-HA immunoprecipitation. Arrows indicate protein bands on silver staining that were analysed by mass spectrometry. c, Purification of recombinant HA–DEPDC5, FLAG–KLHL19 or FLAG–KLHL22 proteins in HEK293T cells. d, CUL3–KLHL22 E3 ligase catalyses DEPDC5 ubiquitination in a cell-free system. Immunopurified HA–DEPDC5 and recombinant wild-type (WT) or mutant (K48R) ubiquitin were incubated with recombinant CUL3–RBX1 and KLHL22 or KLHL19. Asterisk represents HA–DEPDC5. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 3 KLHL22 promotes the ubiquitination of DEPDC5 on multiple lysine residues.
a, Schematic depicting the predicted ubiquitination sites of DEPDC5. b, c, The 5KR mutation prevents K48-linked ubiquitination (b) and degradation (c) of DEPDC5. d, Single mutation of each lysine residue does not impair K48-linked ubiquitination (d) and degradation (e) of DEPDC5. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 4 DEP domain marks the degron of DEPDC5.
a, b, KLHL22 specifically interacts with DEPDC5. c, KLHL22 disassociates from DEPDC5 in response to amino acid starvation. d, Recombinant FLAG–KLHL22 directly interacts with WT but not 5KR HA–DEPDC5 in vitro. e, Schematic depicting the truncations of DEPDC5. f, KLHL22 interacts with DEPDC5 through the DEP domain. g, Deletion of DEP motif prevents KLHL22-mediated degradation of DEPDC5. h, Mutation of each serine, threonine or tyrosine residue in DEP domain to alanine does not affect the interaction between KLHL22 and DEPDC5. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 5 14–3–3 regulates nuclear–cytosolic shuttling of KLHL22.
a, b, Nuclear–cytosolic shuttling of KLHL22 is regulated by amino acids. DAPI, nuclei. Scale bar, 10 μm. c, Schematic depicting the reported phosphorylation sites of KLHL22. d, S18A mutation prevents nuclear accumulation of KLHL22 in amino-acid-deficient conditions. Scale bar, 10 μm. e, Schematic depicting the interaction between FLAG–KLHL22 and 14-3-3 in mass spectrometry analysis using FLAG–KLHL22 as the bait. f, S18A mutation prevents the interaction between KLHL22 and 14-3-3 in amino-acid-deprived conditions. g, KLHL22(S18A) mutant promotes S6K1 phosphorylation in amino-acid-deficient conditions. h, Immunopurification of lysosomes. LAMP2 (lysosome), EEA1 (early endosome), prohibitin (mitochondria), PDI (endoplasmic reticulum) and histone H3 (nucleus). i, Purification of HA–KLHL22. For gel source data, see Supplementary Fig. 1.
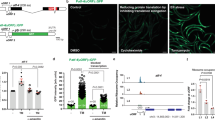
Extended Data Fig. 6 KLHL22 is essential for activation of mTORC1 and downstream events.
a, Schematic depicting CRISPR–Cas9-mediated knockout of KLHL19 or KLHL22 in HEK293T cells. b, KLHL22 is required for amino-acid-dependent lysosomal localization of mTORC1. Scale bar, 10 μm. c, d, Overexpression (c) or depletion (d) of KLHL22 does not affect mTORC1 activity in DEPDC5-deprived cells. e, KLHL22 is required for amino-acid-induced suppression of autophagy. n = 3 biologically independent experiments. f, Depletion of KLHL22 induces autophagy. Percentages of cells with obvious LC3B puncta were calculated. n = 3 biologically independent experiments. ns, no significant difference; ***P < 0.001; two-sided Student’s t-test. For gel source data, see Supplementary Fig. 1.
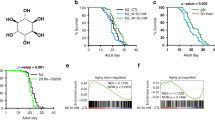
Extended Data Fig. 7 KLHL22 has a conserved role in mice and nematodes.
a, Schematic depicting CRISPR–Cas9-mediated knockout of KLHL19 or KLHL22 in MEF cells. b, Predicted orthologues of KLHL22 in C. elegans. c, Knockdown efficiency of tag-53, mel-26 and T08A11.1. n = 2 biologically independent experiments. d, RNAi targeting T08A11.1 suppresses the developmental delay of worms induced by mel-26 RNAi, but not tag-53 RNAi. n = 3 biologically independent experiments. ns, no significant difference; *P < 0.05; **P < 0.01; ***P < 0.001; two-sided Student’s t-test.
Extended Data Fig. 8 Protein levels of KLHL22 are elevated in human breast cancer.
a, Stable expression of HA–DEPDC5 reduces the size of HEK293T cells. b, Stable expression of FLAG–KLHL22 reverses the reduction in cell size in HEK293T cells stably expressing HA–DEPDC5. c, High expression of FLAG–KLHL22 transforms HEK293T cells. d, According to Oncomine database, mRNA levels of KLHL22 are elevated in breast and prostate cancers and in melanoma. Fold change of KLHL22 transcript levels in tumour tissues (T) to the corresponding normal tissues (NT) are shown. Rank represents KLHL22 rank in ordered list of genes that are upregulated. e, Protein levels of KLHL22 in breast cancer, prostate cancer and melanoma cell lines and corresponding normal cells (MAF10A, PWR1E and HSF). f, mRNA levels of KLHL22 are elevated in tumours of breast cancer patients. T: tumours; ANT: adjacent normal tissues. Quantifications are normalized to actin. n = 3 technically independent experiments. ns, no significant difference; *P < 0.05; **P < 0.01; ***P < 0.001; two-sided Student’s t-test. g, Protein levels of endogenous DEPDC5 are regulated in an amino-acid-sensitive manner in MDA-MB-231 cells. h, Amino acid deprivation or stimulation does not affect mRNA levels of DEPDC5 in MDA-MB-231 cells. Relative mRNA levels of DEPDC5 to GAPDH were quantified using qPCR. n = 3 biologically independent experiments. ns, no significant difference; two-sided Student’s t-test. i, Proteasome inhibitor MG132 increases the protein levels of DEPDC5 in MDA-MB-231 cells. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 9 Deletion of KLHL22 in breast cancer cells prevents mTORC1 activation and tumorigenesis.
a, b, Schematic of CRISPR–Cas9-mediated knockout of KLHL19 or KLHL22 in MDA-MB-231 (a) or MDA-MB-468 (b) breast cancer cells. c, Protein levels of KLHL22 are upregulated in MDA-MB-231/468 cells. d, e, Deletion of KLHL22 in MDA-MB-231 (d) or MDA-MB-468 (e) cells suppresses amino-acid-induced DEPDC5 degradation and S6K1 phosphorylation. f–h, Deletion of KLHL22 suppresses tumour growth of MDA-MB-468 cells. Tumour volume, mean ± s.e.m. (n = 9 mice for WT, n = 8 mice for sgKLHL22_1), ***P < 0.001, two-sided ANOVA (f); tumour images (g); tumour weights 20 days after transplantation, mean ± s.e.m. (n = 6 mice for WT and n = 8 mice for KLHL22 knockouts), ***P < 0.001, two-sided Student’s t-test (h). i–k, Deletion of KLHL22 or treatment with rapamycin suppresses tumour growth of MDA-MB-231 cells. Tumour volume, mean ± s.e.m. (n = 9 mice for sgKLHL22_2 and WT injected with low dose of rapamycin, n = 10 mice for remaining groups), ***P < 0.001, two-sided ANOVA (i); tumour images (j); and tumour weights 20 days after transplantation, mean ± s.e.m. (n = 8 mice for WT and WT injected with low or high dose of rapamycin; n = 6 mice for KLHL22 knockouts), ***P < 0.001, two-sided Student’s t-test (k). For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 10 Proposed model of KLHL22-mediated regulation of mTORC1, ageing and cancer.
In response to amino acids, KLHL22 translocates from the nucleus to the cytosol, where it accumulates on the surface of lysosomes to mediate the ubiquitination and degradation of DEPDC5, an essential subunit of GATOR1. GATOR1 loss of function activates Rag GTPases and subsequently activates mTORC1 (top). KLHL22-mediated DEPDC5 degradation is a conserved mechanism of TORC1 regulation in mammals and nematode. TORC1 hypoactivation due to mel-26 (Ce.KLHL22) depletion extends C. elegans lifespan, whereas TORC1 hyperactivation due to elevated expression of KLHL22 promotes human breast cancer (bottom).
Supplementary information
Supplementary Figure 1
This file contains uncropped scans of the western blots presented in the paper.
Rights and permissions
About this article
Cite this article
Chen, J., Ou, Y., Yang, Y. et al. KLHL22 activates amino-acid-dependent mTORC1 signalling to promote tumorigenesis and ageing. Nature 557, 585–589 (2018). https://doi.org/10.1038/s41586-018-0128-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0128-9
This article is cited by
-
The rapid proximity labeling system PhastID identifies ATP6AP1 as an unconventional GEF for Rheb
Cell Research (2024)
-
Amino acid metabolism in tumor biology and therapy
Cell Death & Disease (2024)
-
CRL2APPBP2-mediated TSPYL2 degradation counteracts human mesenchymal stem cell senescence
Science China Life Sciences (2024)
-
Visualization of breast cancer-related protein synthesis from the perspective of bibliometric analysis
European Journal of Medical Research (2023)
-
Kelch-like proteins in the gastrointestinal tumors
Acta Pharmacologica Sinica (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.