Abstract
Somatic mutations in tet methylcytosine dioxygenase 2 (TET2), which encodes an epigenetic modifier enzyme, drive the development of haematopoietic malignancies1,2,3,4,5,6,7. In both humans and mice, TET2 deficiency leads to increased self-renewal of haematopoietic stem cells with a net developmental bias towards the myeloid lineage1,4,8,9. However, pre-leukaemic myeloproliferation (PMP) occurs in only a fraction of Tet2−/− mice8,9 and humans with TET2 mutations1,3,5,6,7, suggesting that extrinsic non-cell-autonomous factors are required for disease onset. Here we show that bacterial translocation and increased interleukin-6 production, resulting from dysfunction of the small-intestinal barrier, are critical for the development of PMP in mice that lack Tet2 expression in haematopoietic cells. Furthermore, in symptom-free Tet2−/− mice, PMP can be induced by disrupting intestinal barrier integrity, or in response to systemic bacterial stimuli such as the toll-like receptor 2 agonist. PMP was reversed by antibiotic treatment and failed to develop in germ-free Tet2−/− mice, which illustrates the importance of microbial signals in the development of this condition. Our findings demonstrate the requirement for microbial-dependent inflammation in the development of PMP and provide a mechanistic basis for the variation in PMP penetrance observed in Tet2−/− mice. This study will prompt new lines of investigation that may profoundly affect the prevention and management of haematopoietic malignancies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Delhommeau, F. et al. Mutation in TET2 in myeloid cancers. N. Engl. J. Med. 360, 2289–2301 (2009).
Kosmider, O. et al. TET2 gene mutation is a frequent and adverse event in chronic myelomonocytic leukemia. Haematologica 94, 1676–1681 (2009).
Busque, L. et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat. Genet. 44, 1179–1181 (2012).
Abdel-Wahab, O. & Levine, R. L. Mutations in epigenetic modifiers in the pathogenesis and therapy of acute myeloid leukemia. Blood 121, 3563–3572 (2013).
Genovese, G. et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371, 2477–2487 (2014).
Jaiswal, S. et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371, 2488–2498 (2014).
Jan, M., Ebert, B. L. & Jaiswal, S. Clonal hematopoiesis. Semin. Hematol. 54, 43–50 (2017).
Moran-Crusio, K. et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20, 11–24 (2011).
Quivoron, C. et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 20, 25–38 (2011).
Bowman, R. L., Busque, L. & Levine, R. L. Clonal hematopoiesis and evolution to hematopoietic malignancies. Cell Stem Cell 22, 157–170 (2018).
Kunimoto, H. et al. Cooperative epigenetic remodeling by TET2 loss and NRAS mutation drives myeloid transformation and MEK inhibitor sensitivity. Cancer Cell 33, 44–59.e48 (2018).
Kogan, S. C. et al. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood 100, 238–245 (2002).
Ko, M. et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc. Natl Acad. Sci. USA 108, 14566–14571 (2011).
Li, Z. et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood 118, 4509–4518 (2011).
Boettcher, S. & Manz, M. G. Regulation of inflammation- and infection-driven hematopoiesis. Trends Immunol. 38, 345–357 (2017).
Sartor, R. B. Microbial influences in inflammatory bowel diseases. Gastroenterology 134, 577–594 (2008).
Marchiando, A. M., Graham, W. V. & Turner, J. R. Epithelial barriers in homeostasis and disease. Annu. Rev. Pathol. 5, 119–144 (2010).
Chassaing, B., Aitken, J. D., Malleshappa, M. & Vijay-Kumar, M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr. Protoc. Immunol. 104, 15.25.1–15.25.14 (2014).
Shen, L., Weber, C. R., Raleigh, D. R., Yu, D. & Turner, J. R. Tight junction pore and leak pathways: a dynamic duo. Annu. Rev. Physiol. 73, 283–309 (2011).
Chapman, C. G. et al. TET-catalyzed 5-hydroxymethylcytosine regulates gene expression in differentiating colonocytes and colon cancer. Sci. Rep. 5, 17568 (2015).
Zhao, Z. et al. The catalytic activity of TET2 is essential for its myeloid malignancy-suppressive function in hematopoietic stem/progenitor cells. Leukemia 30, 1784–1788 (2016).
Eren, A. M. et al. Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol. Evol. 4, 1111–1119 (2013).
Wells, J. M. Immunomodulatory mechanisms of lactobacilli. Microb. Cell Fact. 10, S17 (2011).
Kristinsson, S. Y. et al. Chronic immune stimulation might act as a trigger for the development of acute myeloid leukemia or myelodysplastic syndromes. J. Clin. Oncol. 29, 2897–2903 (2011).
Panteli, K. E. et al. Serum interleukin (IL)-1, IL-2, sIL-2Ra, IL-6 and thrombopoietin levels in patients with chronic myeloproliferative diseases. Br. J. Haematol. 130, 709–715 (2005).
Miller, C. L. & Lai, B. Human and mouse hematopoietic colony-forming cell assays. Methods Mol. Biol. 290, 71–89 (2005).
Johnson, D. E., O’Keefe, R. A. & Grandis, J. R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 15, 234–248 (2018).
Cimmino, L. et al. Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. Cell 170, 1079–1095.e1020 (2017).
Slack, E. et al. Innate and adaptive immunity cooperate flexibly to maintain host–microbiota mutualism. Science 325, 617–620 (2009).
Meisel, M. et al. Interleukin-15 promotes intestinal dysbiosis with butyrate deficiency associated with increased susceptibility to colitis. ISME J. 11, 15–30 (2017).
Stefka, A. T. et al. Commensal bacteria protect against food allergen sensitization. Proc. Natl Acad. Sci. USA 111, 13145–13150 (2014).
Furuta, G. T. et al. Hypoxia-inducible factor 1-dependent induction of intestinal trefoil factor protects barrier function during hypoxia. J. Exp. Med. 193, 1027–1034 (2001).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Johnson, W. E., Li, C. & Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007).
Storey, J. D., Bass, A. J., Dabney, A. & Robinson, D. qvalue: Q-value estimation for false discovery rate control. R package version 2.8.0. http://github.com/jdstorey/qvalue (2015).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Huse, S. M. et al. Comparison of brush and biopsy sampling methods of the ileal pouch for assessment of mucosa-associated microbiota of human subjects. Microbiome 2, 5 (2014).
Eren, A. M., Vineis, J. H., Morrison, H. G. & Sogin, M. L. A filtering method to generate high quality short reads using illumina paired-end technology. PLoS ONE 8, e66643 (2013).
Eren, A. M. et al. Minimum entropy decomposition: unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J. 9, 968–979 (2015).
Huse, S. M. et al. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 4, e1000255 (2008).
Oksanen, J., Kindt, R., Legendre, P., O’Hara, B. & Stevens, M. H. Vegan: community ecology package. https://CRAN.R-project.org/package=vegan (2007).
Cummings, R. J. et al. Different tissue phagocytes sample apoptotic cells to direct distinct homeostasis programs. Nature 539, 565–569 (2016).
Acknowledgements
We thank the University of Chicago DNA Sequencing Core facility for assistance with sequencing; the Calcul Québec and Compute Canada for providing access to the supercomputer Briaree from the University of Montreal; the Histology Core facility at the University of Chicago Human Tissue Resource Center for assistance with histology; S. Hwang (University of Chicago) for providing LysM-Cre mice; and V. Abadie (Université de Montréal) for discussions and critical reading of the manuscript. This work was supported by grants from the Cancer Center Support Grant P30CA014599 to B.J.; Digestive Diseases Research Core Center P30DK42086 at the University of Chicago to R.H., E.B.C., C.R.W. and B.J.; P01DK072201, R01DK110352 and 5R01CA161373 to S.A.L. and G.C.F.; F32 DK105728-01A1 to J.F.P.; CCFA Research Fellowship Award (ID: 480735) to M.M.; FWF Austrian Science Fund (P30324-B21) and Christian Doppler Society (I-CARE) to G.B.; and by a CIHR grant MOP (#20003029), a Canada Research Chair to E.F.V. and a Canada Research Chair to L.B.B. A.P. was supported by a fellowship from FRQS.
Reviewer information
Nature thanks S. Grivennikov, K. Shannon, Y. Xiong and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
M.M., R.H. and B.J. conceived and designed the research, guided the interpretation of the results and wrote the manuscript. M.M. and R.H. performed the majority of the experiments and data analysis. A.P. and L.B.B. performed RNA sequencing experiments and data analysis. L.C. helped with various experiments. Z.M.E. and D.L.R. performed bacterial cultures and species identification. T.M. performed cell-sorting experiments. J.F.P. performed histology experiments. J.D.E. and M.W.M. helped with intestinal permeability assays. H.J.G. helped with germ-free mouse experiments. N.T. performed cytokine measurements. R.B. performed RT-PCR experiments. M.B. performed 16S PCR experiments. Y.W., Y.L. and C.R.W. performed ZO-1 immunofluorescence experiments and analysis. B.D.M. helped with flow cytometry experiments. V.D., S.M.K. and M.A.Z. helped with intestinal tissue sample collection. G.C.F. performed TUNEL and caspase 3 experiments and analysis. A.M.E. performed 16S analysis. S.A.L., G.B., E.B.C., A.M.E., C.R.W., L.B., L.A.G., E.F.V. and L.B.B. provided critical intellectual input and technical support. T.M., L.A.G., L.B.B. and B.J. critically edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Tet2−/− mice display variability in PMP penetrance that correlates with bacteraemia.
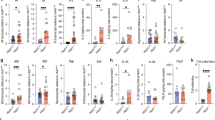
a, Penetrance of PMP assessed by splenomegaly, splenic LSK cell expansion and CD11b+Gr1+ myeloid cell expansion in spleen and peripheral blood. b–e, Correlation between frequency of peripheral-blood CD11b+Gr1+ myeloid cells (peripheral-blood biomarker) and numbers of CD11b+Gr1+ myeloid cells in spleen (b) and peripheral blood (c), numbers of LSK cells (d) and splenomegaly (e), in Tet2+/+ and Tet2−/− mice. In b–e, dotted vertical lines indicate level of 16% for peripheral-blood CD11b+Gr1+ myeloid cells (level determined by the distribution in wild-type littermates). The peripheral-blood biomarker (frequency of CD11b+Gr1+ myeloid cells) was used to categorize Tet2−/− mice either as symptom free (≤16% CD11b+Gr1+ myeloid cells) or as mice with PMP (>16% CD11b+Gr1+ myeloid cells). Spearman correlation test excluding Tet2+/+ mice. Red lines indicate regression line calculated for Tet2−/− data points (n = 10 (blue), 23 mice (red). f–i, Symptom-free, Tet2−/− mice with PMP and littermate controls were used. f, Representative dot blots are shown of LSK cells in the spleen, CD11b+Gr1+ myeloid cells in the spleen and peripheral blood, and lymphocytes in peripheral blood. Data are representative of five independent experiments with similar results. g, Spleen weight (top; n = 11 (blue), 9 (red, PMP) or 5 mice (red, symptom free)) and numbers of CD11b+Gr1+ myeloid cells in the spleen (bottom; n = 10 (blue), 12 (red, PMP) or 7 mice (red, symptom free)). h, Numbers per ml of peripheral blood of live CD45+ gated white blood cells (WBC) (top left), lymphocytes (top right), CD11b+ monocytes (bottom left) and CD11b+Gr1+ myeloid cells (bottom right) (n = 15 (blue), 15 (red, PMP) or 32 mice (red, symptom free)). i, In vitro HSC self-renewal colony-forming assay of haematopoietic progenitors of the bone marrow (n = 3 mice). Mean ± s.e.m. j, k, Correlation between 16S gene copies in the peripheral blood and spleen weight (n = 16 mice) (j) or numbers of CD11b+ monocytes (k top left), percentage of lymphocytes (k top right), numbers of lymphocytes (k bottom left), and numbers of leukocytes (WBC) (k bottom right) in the peripheral blood (n = 40 mice). Pearson correlation test. l, Bacterial colonies from Fig. 1e, f identified by 16S sequencing; blue rectangles indicate presence of bacteria. In g, h, boxes represent median values and interquartile ranges; whiskers represent minimum and maximum values. One-way ANOVA, Sidak’s post hoc test. Data are representative of at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Fig. 2 Tet2−/− mice display no anatomical changes in the jejunum.
a, Representative images of haematoxylin and eosin-stained sections of the jejunum. b, c, Immunofluorescence analyses of sections of the jejunum of Tet2+/+ and Tet2−/− mice. b, No difference was observed between the groups in the number of apoptotic cells using the TUNEL assay (red). Nuclei of intestinal cells were visualized with DAPI (blue). c, No difference was observed in the number of apoptotic (cleaved caspase 3-positive (CC3), red) epithelial cells (pan-keratin-positive, (PanK), green) between the groups. In b, c, Jejunum sections of diphtheria-toxin-treated VDTR mice were used as a positive control. Data are representative of at least two independent experiments.
Extended Data Fig. 3 Disruption of barrier integrity drives PMP.
a, FITC–dextran concentration in blood plasma correlates with numbers of LSK cells (left, n = 12 mice), frequency of CD11b+Gr1+ myeloid cells in the spleen (middle left, n = 10 mice), peripheral blood (middle right, n = 15 mice) and frequency of lymphocytes (right, n = 15 mice). Pearson correlation test. b, Schematic of DSS treatment of symptom-free Tet2f/fVavcre mice that are over 20 weeks old, and littermate controls. c, Numbers of CD11b+ monocytes (left), lymphocytes (middle) and leukocytes (WBC) (right) before, during and after DSS treatment (n = 5 (blue) or 8 (orange) mice). Mean ± s.e.m. d, Percentage of CD11b+Gr1+ myeloid cells (left) and numbers of GMP (right) at end point analysis (n = 5 (Tet2f/fcre−, without DSS), 4 (Tet2f/fVavcre, without DSS), 6 (Tet2f/fcre−, with DSS) or 8 (Tet2f/fVavcre, with DSS) mice). Centre is median, Kruskal–Wallis, Dunn’s post hoc test. *P < 0.05, **P < 0.01. Data are representative of at least two independent experiments.
Extended Data Fig. 4 Tight junction ZO-1 is markedly reduced in the jejunum but not in the colon of Tet2−/− mice.
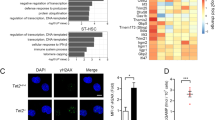
a, RNA sequencing of whole intestinal tissue of duodenum, jejunum, ileum and colon of three Tet2−/− and three Tet2+/+ mice. Venn diagram illustrating the number of differentially expressed genes (|log2(fold change)| > 0.5 and false discovery rate < 0.1); see Supplementary Table in Tet2−/− mice compared to littermate controls. b, RNA sequencing of the jejunum of seven Tet2−/− and seven Tet2+/+ mice (the full list of differentially expressed genes is shown in Supplementary Table. Heat map of a selection of antimicrobial peptides and tight junction genes. c, Gene expression of tight junction gene (ZO-1, Tjp1) in the jejunum (left; n = 12 (Tet2+/+) or 9 (Tet2−/−) mice) and colon (right; n = 9 mice in both cases). Centre is mean, two-tailed unpaired t-test. d, ZO-1 immunofluorescence shows reduced tight junction staining of epithelial cells in the jejunum (left) but no difference in colonic epithelial cells (right) of Tet2−/− mice. Representative images are shown from Tet2+/+ (n = 3 mice) and Tet2−/− mice (n = 7 mice). Scale bars, 100 μm. Green, ZO-1; blue, DAPI. **P < 0.01. Data are representative of at least two independent experiments.
Extended Data Fig. 5 Absence of Tet2 expression in haematopoietic cells leads to barrier dysfunction and PMP.
a, Intestinal permeability measurement by blood plasma FITC–dextran concentrations (n = 16 (Tet2f/fcre−), 6 (Tet2f/fVavcre), 12 (Tet2f/fVillincre) or 12 (Tet2f/fLysMcre) mice). b, c, Gene expression of antimicrobial peptides Retlnb (b left), Ang4 (b right) and (c) tight junction genes ZO-1 (Tjp1) (left), occludin (Ocln) (middle), and desmoplakin (Dsp) (right) in the jejunum (n = 13 (Tet2f/fcre−), 7 (Tet2f/fVavcre), 9 (Tet2f/fVillincre) or 10 (Tet2f/fLysMcre) mice). d, Representative dot blots (left) and numbers of LSK cells (right) in the spleen, CD11b+Gr1+ myeloid cells in the spleen (n = 6 (Tet2f/fcre−), 12 (Tet2f/fVavcre), 10 (Tet2f/fVillincre) or 10 (Tet2f/fLysMcre) mice) and peripheral blood, and lymphocytes in the peripheral blood (n = 10 (Tet2f/fcre−), 8 (Tet2f/fVavcre), 10 (Tet2f/fVillincre) or 10 (Tet2f/fLysMcre) mice). e, White blood cell (WBC) count (top) and cell number per ml of blood of CD11b+ monocytes (bottom) (n = 10 (Tet2f/fcre−), 8 (Tet2f/fVavcre), 10 (Tet2f/fVillincre) or 10(Tet2f/fLysMcre) mice). f, Spleen weight is shown (n = 10 (Tet2f/fcre−), 6 (Tet2f/fVavcre), 10 (Tet2f/fVillincre) or 10 (Tet2f/fLysMcre) mice). In a–f, centre is mean, one-way ANOVA, Sidak’s post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Data are representative of three independent experiments.
Extended Data Fig. 6 Microbial signals are sufficient for PMP in Tet2−/− mice.
a, TLR2 activation measured using a HEK-TLR2 reporter assay (n = 5 biological replicates). b, Schematic of Pam3CSK4 in vivo treatment of symptom-free Tet2f/fVavcre mice that are over 20 weeks old, and littermate controls. c, Numbers of CD11b+ monocytes prior (day 0), during treatment (day 2) and at end point analysis (day 14) (n = 6 mice). Mean ± s.e.m. d, Percentage of CD11b+Gr1+ myeloid cells (left, n = 5 (Tet2f/fcre−, no Pam3CSK4), 6 (Tet2f/fcre−, with Pam3CSK4), 7 (Tet2f/fVavcre, no Pam3CSK4) or 6 (Tet2f/fVavcre, with Pam3CSK4) mice) and numbers of GMP cells (right, n = 6 (Tet2f/fcre−, no Pam3CSK4), 6 (Tet2f/fcre−, with Pam3CSK4), 7 (Tet2f/fVavcre, no Pam3CSK4) or 6 (Tet2f/fVavcre, with Pam3CSK4) mice). e, Intestinal permeability measurement by blood plasma FITC–dextran concentrations at end point analysis (n = 6 mice in all cases). a, c–e, Centre is mean. a, d, One-way ANOVA, Sidak’s post hoc test, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Data are representative of at least two independent experiments.
Extended Data Fig. 7 Microbial community structures are similar between Tet2−/− mice and littermate controls.
a, b, Non-metric multidimensional scaling of samples based on their oligotype profiles (with Bray–Curtis distance). a, Microbial community structures within the same body site—that is, jejunum (left, n = 14 (Tet2−/− (KO)) or 9 mice (Tet2+/+(WT))), colon (middle, n = 7 (KO) or 5 mice (WT)) and faeces (right, n = 6 (KO) or 5 mice (WT))—do not differ significantly across genotypes based on envfit test with 999 permutations (P > 0.05, two-tailed). b, Microbial community structures within the same genotype—WT (top, n = 5 (colonic sample (COL) or faecal sample (FEC)) or 9 mice (jejunal sample (JEJ)) and KO (bottom, n = 6 (FEC), 7 (COL) or 14 mice (JEJ))—significantly differ across body sites based on the same test (P < 0.001). c, d, Heat-map displays of oligotypes across samples. Clustering dendrograms are computed with Bray–Curtis distance and average linkage algorithm using the oligotype profiles. The intensity of colours indicates the per cent abundance of a given oligotype (rows) in a given sample (columns). c, Comparison of genotypes for samples collected from a single body site. d, Comparison of genotypes for samples collected across body sites. e, Co-housing does not affect the development of PMP in Tet2f/fVavcre mice. Representative cage configurations of 3 litters from Tet2f/fVavcre mice with PMP that are over 20 weeks old (red), symptom-free Tet2f/fVavcre mice (grey), and littermate controls (blue). CON, luminal content; SCR, scraping samples.
Extended Data Fig. 8 Microbial signals are required for PMP in Tet2−/− mice.
a, b, Germ-free, SPF-housed Tet2−/− mice that are over 40 weeks old, and germ-free wild-type controls, analysed for percentage of lymphocytes (a left), numbers of lymphocytes (a middle left), numbers of CD11b+ monocytes (a middle right) and numbers of leukocytes (WBC) (a right) (n = 6 (Tet2+/+, GF), 7 (Tet2−/−, GF) or 7 (Tet2−/−, SPF) mice), and percentage of CD11b+Gr1+ myeloid cells (b left) and numbers of GMP cells (b right), (n = 7 (Tet2+/+, GF), 4 (Tet2−/−, GF) or 5 (Tet2−/−, SPF) mice). c, Mice treated with antibiotics (ABX) before onset of PMP (see schematic in Fig. 3c. Numbers of CD11b+Gr1+ myeloid cells (left; n = 14 (Tet2+/+, no ABX), 11 (Tet2−/−, no ABX), 7 (Tet2+/+, with ABX) or 7 (Tet2−/−, with ABX) mice) and LSK cells (right; n = 12 (Tet2+/+, no ABX), 8 (Tet2−/−, no ABX), 7 (Tet2+/+, with ABX) or 7 (Tet2−/−, with ABX) mice). d, Tet2−/− mice monitored for the number of CD11b+ monocytes (left, n = 7 mice) and 16S gene copies in the faeces (right, n = 6 mice) before, during and after antibiotics treatment. Mean ± s.e.m., repeated measures one-way ANOVA, Sidak’s post hoc test. In a–c, centre is mean, one-way ANOVA, Sidak’s post hoc test, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Data are representative of at least two independent experiments.
Extended Data Fig. 9 Antibiotic treatment reverses PMP in Tet2−/− mice.
a–j, Tet2−/− mice with PMP that are over 20 weeks old, and littermate controls, were treated with and without antibiotics (ABX) for four weeks. a, Numbers of peripheral-blood CD11b+ monocytes (n = 13 mice). Lines connect values obtained from the same mouse sampled before (−) and after (+) ABX treatment. b, c, Representative dot blots and percentage of CD11b+Gr1+ myeloid cells (b) (n = 20 (Tet2+/+, no ABX), 21 (Tet2−/−, no ABX), 6 (Tet2+/+, with ABX) or 14 (Tet2−/−, with ABX) mice) and LSK cells in the spleen (c) (n = 11(Tet2+/+, no ABX), 12 (Tet2−/−, no ABX), 6 (Tet2+/+, with ABX) or 12 (Tet2−/−, with ABX) mice). d, Representative dot blots and numbers of bone marrow-derived LSK cells (n = 11(Tet2+/+, no ABX), 13 (Tet2−/−, no ABX), 6 (Tet2+/+, with ABX) or 12 (Tet2−/−, with ABX) mice). e, f, Representative dot blots and numbers of splenic (e) (n = 6 (Tet2+/+, no ABX), 12 (Tet2−/−, no ABX), 5 (Tet2+/+, with ABX) or 12 (Tet2−/−, with ABX) mice) and bone marrow-derived (f) LSK gated CD34+Flt3− (Flt3 is also known as CD135) short-term (ST)-HSCs and CD34−Flt3− long-term (LT)-HSCs (n = 6 (Tet2+/+, no ABX), 13 (Tet2−/−, no ABX), 5 (Tet2+/+, with ABX) or 12 (Tet2−/−, with ABX) mice). g, h, Representative dot blots and numbers of splenic (g) (n = 5(Tet2+/+, no ABX), 10 (Tet2−/−, no ABX), 5 (Tet2+/+, with ABX) or 10 (Tet2−/−, with ABX) mice) and bone marrow-derived (h) LSK gated CD150+ CD48− cells (n = 5 (Tet2+/+, no ABX), 11 (Tet2−/−, no ABX), 5 (Tet2+/+, with ABX) or 10 (Tet2−/−, with ABX) mice). i, j, Representative dot blots and numbers of splenic (i) (n = 6 (Tet2+/+, no ABX), 12 (Tet2−/−, no ABX), 5 (Tet2+/+, with ABX) or 12 (Tet2−/−, with ABX) mice) and bone marrow-derived (j) c-Kit+Sca-1− (LK gated) GMP, common myeloid progenitor (CMP) and megakaryocyte–erythroid progenitor (MEP) cells (n = 6 (Tet2+/+, no ABX), 13 (Tet2−/−, no ABX), 5 (Tet2+/+, with ABX) or 12 (Tet2−/−, with ABX) mice). In b–j, centre is mean, one-way ANOVA, Sidak’s post hoc test. Data are representative of at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Fig. 10 Bacteria-induced IL-6 is required for PMP in Tet2−/− mice.
a, Gene expression of Il6 in the spleen of SPF-housed Tet2−/− mice, littermates and germ-free Tet2−/− mice (n = 7 (Tet2+/+, SPF), 10 (Tet2−/−, SPF) or 4 (Tet2−/−, GF) mice). b, IL-6 cytokine levels in blood plasma of Tet2−/− mice with PMP treated with antibiotics (ABX) for four weeks (n = 5 mice). Lines connect values obtained from the same mouse sampled before and after antibiotics treatment. Two-tailed paired t-test. c, IL-6 cytokine levels in blood plasma of DSS-treated symptom-free Tet2f/fVavcre mice that are over 20 weeks old, and littermate controls (see schematic in Extended Data Fig. 3b (n = 6 (Tet2+/+) or 9 (Tet2−/−) mice). d, IL-6 cytokine levels in blood plasma of Tet2f/fVavcre mice that are over 20 weeks old, and littermate controls, treated with the TLR1/2 agonist Pam3CSK4 (see schematic in Extended Data Fig. 6b (n = 6 (Tet2+/+) or 8 (Tet2−/−) mice). e, Correlation between IL-6 cytokine levels in blood plasma of Tet2−/− mice and numbers of peripheral-blood CD11b+ monocytes (n = 49 mice). Pearson correlation test. f, g, In vitro HSC self-renewal colony-forming assay of haematopoietic progenitors of the spleen from Tet2−/− mice (red lines) and littermate controls (blue lines) in the presence of anti-IL-6 antibody or isotype control (ISO) after the first replating (n = 3 mice). g, Representative images of colonies after the 5th replating. Scale bars, 100 μm. h, Representative histogram (left) and quantification of (right) mean fluorescence intensity (MFI) of IL-6Rα+c-Kit+Sca-1− (LK gated) CD34+FcγRIII/II+ GMPs from the bone marrow, (n = 5 (Tet2+/+, SPF), 6 (Tet2−/−, SPF), 4 (Tet2+/+, GF) or 3 (Tet2−/−, GF) mice). i, Representative flow cytometry plot of Stat3 phosphorylation (pY705) response after 30-min stimulation with 10 ng ml−1 IL-6 in splenic c-Kit+Sca-1− (LK gated) CD34+ and CD34− myeloid progenitors (MP). j, Numbers of CD11b+ myeloid cells (n = 7 (Tet2+/+) or 6 (Tet2−/−) mice). k, l, CD11b+F4/80+ macrophages and CD11b+Gr1+ myeloid cells of the spleen, from Tet2−/− mice and littermate controls, were FACS sorted. Gene expression of Il6 in macrophages (k) and CD11b+Gr1+ cells (l) (n = 7 (Tet2+/+) or 6 (Tet2−/−) mice). Centre is median. m, Representative histogram (left) and quantification of (right) MFI of IL-6Rα+CD11b+Gr1+ myeloid cells in the spleen (n = 5 (Tet2+/+, SPF), 6 (Tet2−/−, SPF), 4 (Tet2+/+, GF) or 4 (Tet2−/−, GF) mice). n, Representative histogram (left) and quantification of (right) MFI of IL-6Rα+c-Kit+Sca-1− (LK gated) CD34+FcγRIII/II+ GMPs from the spleen of Tet2f/fLysMcre mice and littermate controls (n = 5 mice). Centre is mean. o, p, anti-IL-6 antibody (+) or ISO treatment (−) of Tet2−/− mice with PMP that are over 20 weeks old, and littermate controls (see schematic in Fig. 4g. o, Percentage of CD11b+Gr1+ myeloid cells (left; n = 10 (Tet2+/+, with anti-IL-6), 6 (Tet2−/−, with anti-IL-6) or 7 (Tet2−/−, without anti-IL-6) mice) and numbers of GMPs (right; n = 11 (Tet2+/+, with anti-IL-6), 6 (Tet2−/−, with anti-IL-6) or 8 (Tet2−/−, without anti-IL-6) mice) in the spleen. p, Intestinal permeability was assessed by blood plasma FITC–dextran concentrations (n = 6 mice in all cases). q, IL-6 in supernatants from intestinal explants: jejunum (left; n = 6 (Tet2+/+) or 5 (Tet2−/−) mice) and colon (right; n = 6 mice in both cases). Centre is mean. a, h, m, p, Centre is mean, one-way ANOVA, Sidak’s post hoc test. c, d, j, Centre is median, two-tailed Mann–Whitney U-test. o, Centre is median, Kruskal–Wallis, Dunn’s post hoc test. Data are representative of at least three independent experiments; *P < 0.05, **P < 0.01, ***P < 0.001. r, Model showing that extrinsic (microbial-induced inflammatory) and intrinsic (IL-6Rα expression) signals are required for PMP in Tet2−/− mice. In detail, small-intestinal barrier dysfunction (reduced ZO-1 and upregulation of defence response genes), which occurs spontaneously or upon intestinal damage, leads to bacterial translocation and to high levels of IL-6. Bacterial translocation can be bypassed when Tet2-deficient mice receive systemic microbial signals. Microbial-induced IL-6 is sensed by Tet2−/− myeloid progenitor (MP) cells that overexpress IL-6Rα and are highly sensitive to IL-6 (Stat3 (pY705)). Subsequently, MPs expand upon IL-6 signals and preferentially differentiate into mature myeloid cells with IL-6-producing capacities. This cycle results in the development of PMP. Treatment with antibiotics or neutralizing anti-IL-6 antibody can revert PMP, indicating that microbial inflammatory signals are required for PMP in the context of Tet2 deficiency. However, whether bacteria-induced inflammatory signals also create a permissive environment that induces the acquisition of cooperative oncogenic mutations that lead to the development of leukaemia remains to be determined. The mechanisms through which Tet2 deficiency in haematopoietic cells leads to a microbiota-dependent impairment of gut barrier function remains to be addressed.
Supplementary information
Supplementary Figure 1
Flow cytometry gating strategy
Supplementary Table 1
RNA-seq mapping statistics
Supplementary Table 2
A list of protein-coding genes ranked by statistical evidence of differential expression in Tet2−/− mice (Duodenum, Jejunum, Ileum, Colon)
Supplementary Table 3
A list of protein-coding genes ranked by statistical evidence of differential expression in Tet2−/− mice jejunum (7 samples)
Supplementary Table 4
16S rRNA Matrix counts
Rights and permissions
About this article
Cite this article
Meisel, M., Hinterleitner, R., Pacis, A. et al. Microbial signals drive pre-leukaemic myeloproliferation in a Tet2-deficient host. Nature 557, 580–584 (2018). https://doi.org/10.1038/s41586-018-0125-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0125-z
This article is cited by
-
Made to order: emergency myelopoiesis and demand-adapted innate immune cell production
Nature Reviews Immunology (2024)
-
Tissue mosaicism following stem cell aging: blood as an exemplar
Nature Aging (2024)
-
Robustness of cancer microbiome signals over a broad range of methodological variation
Oncogene (2024)
-
Triangulating nutrigenomics, metabolomics and microbiomics toward personalized nutrition and healthy living
Human Genomics (2023)
-
Intestinal barrier functions in hematologic and oncologic diseases
Journal of Translational Medicine (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.