Abstract
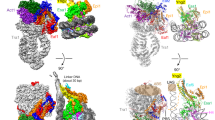
In the eukaryotic nucleus, DNA is packaged in the form of nucleosomes, each of which comprises about 147 base pairs of DNA wrapped around a histone protein octamer. The position and histone composition of nucleosomes is governed by ATP-dependent chromatin remodellers1,2,3 such as the 15-subunit INO80 complex4. INO80 regulates gene expression, DNA repair and replication by sliding nucleosomes, the exchange of histone H2A.Z with H2A, and the positioning of + 1 and −1 nucleosomes at promoter DNA5,6,7,8. The structures and mechanisms of these remodelling reactions are currently unknown. Here we report the cryo-electron microscopy structure of the evolutionarily conserved core of the INO80 complex from the fungus Chaetomium thermophilum bound to a nucleosome, at a global resolution of 4.3 Å and with major parts at 3.7 Å. The INO80 core cradles one entire gyre of the nucleosome through multivalent DNA and histone contacts. An Rvb1/Rvb2 AAA+ ATPase heterohexamer is an assembly scaffold for the complex and acts as a ‘stator’ for the motor and nucleosome-gripping subunits. The Swi2/Snf2 ATPase motor binds to nucleosomal DNA at superhelical location −6, unwraps approximately 15 base pairs, disrupts the H2A–DNA contacts and is poised to pump entry DNA into the nucleosome. Arp5 and Ies6 bind superhelical locations −2 and −3 to act as a counter grip for the motor, on the other side of the H2A–H2B dimer. The Arp5 insertion domain forms a grappler element that binds the nucleosome dyad, connects the Arp5 actin-fold and entry DNA over a distance of about 90 Å and packs against histone H2A–H2B near the ‘acidic patch’. Our structure together with biochemical data8 suggests a unified mechanism for nucleosome sliding and histone editing by INO80. The motor is part of a macromolecular ratchet, persistently pumping entry DNA across the H2A–H2B dimer against the Arp5 grip until a large nucleosome translocation step occurs. The transient exposure of H2A–H2B by motor activity as well as differential recognition of H2A.Z and H2A may regulate histone exchange.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bartholomew, B. Regulating the chromatin landscape: structural and mechanistic perspectives. Annu. Rev. Biochem. 83, 671–696 (2014).
Narlikar, G. J., Sundaramoorthy, R. & Owen-Hughes, T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 154, 490–503 (2013).
Clapier, C. R., Iwasa, J., Cairns, B. R. & Peterson, C. L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 18, 407–422 (2017).
Shen, X., Mizuguchi, G., Hamiche, A. & Wu, C. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406, 541–544 (2000).
Papamichos-Chronakis, M., Watanabe, S., Rando, O. J. & Peterson, C. L. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 144, 200–213 (2011).
Udugama, M., Sabri, A. & Bartholomew, B. The INO80 ATP-dependent chromatin remodeling complex is a nucleosome spacing factor. Mol. Cell. Biol. 31, 662–673 (2011).
Krietenstein, N. et al. Genomic nucleosome organization reconstituted with pure proteins. Cell 167, 709–72.e712 (2016).
Brahma, S. et al. INO80 exchanges H2A.Z for H2A by translocating on DNA proximal to histone dimers. Nat. Commun. 8, 15616 (2017).
Jiang, C. & Pugh, B. F. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 10, 161–172 (2009).
Liu, X., Li, M., Xia, X., Li, X. & Chen, Z. Mechanism of chromatin remodelling revealed by the Snf2–nucleosome structure. Nature 544, 440–445 (2017).
Sundaramoorthy, R. et al. Structural reorganization of the chromatin remodeling enzyme Chd1 upon engagement with nucleosomes. eLife 6, e22510 (2017).
Farnung, L., Vos, S. M., Wigge, C. & Cramer, P. Nucleosome–Chd1 structure and implications for chromatin remodelling. Nature 550, 539–542 (2017).
Tosi, A. et al. Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex. Cell 154, 1207–1219 (2013).
Chen, L. et al. Subunit organization of the human INO80 chromatin remodeling complex: an evolutionarily conserved core complex catalyzes ATP-dependent nucleosome remodeling. J. Biol. Chem. 286, 11283–11289 (2011).
Nguyen, V. Q. et al. Molecular architecture of the ATP-dependent chromatin-remodeling complex SWR1. Cell 154, 1220–1231 (2013).
Mizuguchi, G. et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348 (2004).
Albert, I. et al. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446, 572–576 (2007).
Willhoft, O., Bythell-Douglas, R., McCormack, E. A. & Wigley, D. B. Synergy and antagonism in regulation of recombinant human INO80 chromatin remodeling complex. Nucleic Acids Res. 44, 8179–8188 (2016).
Chen, L., Conaway, R. C. & Conaway, J. W. Multiple modes of regulation of the human Ino80 SNF2 ATPase by subunits of the INO80 chromatin-remodeling complex. Proc. Natl Acad. Sci. USA 110, 20497–20502 (2013).
Yen, K., Vinayachandran, V. & Pugh, B. F. SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell 154, 1246–1256 (2013).
Gangaraju, V. K. & Bartholomew, B. Dependency of ISW1a chromatin remodeling on extranucleosomal DNA. Mol. Cell. Biol. 27, 3217–3225 (2007).
Zhou, C. Y. et al. Regulation of Rvb1/Rvb2 by a domain within the INO80 chromatin remodeling complex implicates the yeast Rvbs as protein assembly chaperones. Cell Reports 19, 2033–2044 (2017).
Aramayo, R. J. et al. Cryo-EM structures of the human INO80 chromatin-remodeling complex. Nat. Struct. Mol. Biol. 25, 37–44 (2017).
Koldewey, P., Horowitz, S. & Bardwell, J. C. A. Chaperone–client interactions: non-specificity engenders multifunctionality. J. Biol. Chem. 292, 12010–12017 (2017).
Lakomek, K., Stoehr, G., Tosi, A., Schmailzl, M. & Hopfner, K. P. Structural basis for dodecameric assembly states and conformational plasticity of the full-length AAA + ATPases Rvb1•Rvb2. Structure 23, 483–495 (2015).
Rivera-Calzada, A. et al. The structure of the R2TP complex defines a platform for recruiting diverse client proteins to the HSP90 molecular chaperone system. Structure 25, 1145–1152.e4 (2017).
Saha, A., Wittmeyer, J. & Cairns, B. R. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 16, 2120–2134 (2002).
Dürr, H., Körner, C., Müller, M., Hickmann, V. & Hopfner, K. P. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell 121, 363–373 (2005).
Willhoft, O. et al. Crosstalk within a functional INO80 complex dimer regulates nucleosome sliding. eLife 6, e25782 (2017).
Zhou, C. Y. et al. The yeast INO80 complex operates as a tunable DNA length-sensitive switch to regulate nucleosome sliding. Mol. Cell 69, 677–688.e9 (2018).
Watanabe, S., Radman-Livaja, M., Rando, O. J. & Peterson, C. L. A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme. Science 340, 195–199 (2013).
Fitzgerald, D. J. et al. Protein complex expression by using multigene baculoviral vectors. Nat. Methods 3, 1021–1032 (2006).
Klinker, H., Haas, C., Harrer, N., Becker, P. B. & Mueller-Planitz, F. Rapid purification of recombinant histones. PLoS ONE 9, e104029 (2014).
Dyer, P. N. et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2004).
Lowary, P. T. & Widom, J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 (1998).
Levendosky, R. F., Sabantsev, A., Deindl, S. & Bowman, G. D. The Chd1 chromatin remodeler shifts hexasomes unidirectionally. eLife 5, e21356 (2016).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Biasini, M. et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 (2014).
Yang, J. et al. The I-TASSER suite: protein structure and function prediction. Nat. Methods 12, 7–8 (2015).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Gerber, P. R. & Müller, K. MAB, a generally applicable molecular force field for structure modelling in medicinal chemistry. J. Comput. Aided Mol. Des. 9, 251–268 (1995).
Qi, Y. et al. CHARMM-GUI MDFF/xMDFF utilizer for molecular dynamics flexible fitting simulations in various environments. J. Phys. Chem. B 121, 3718–3723 (2017).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2017).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Acknowledgements
We thank P. Korber and E. Oberbeckmann for help with biochemical analysis of INO80, E. Hurt for help with C. thermophilum genome annotations, O. Berninghausen for help with data collection, and K. Lammens, K. Knoll, R. Byrne, G. Stöhr and G. Witte for discussions and technical help. We thank the LMU Protein Analysis Unit and the MPI of Biochemistry cryo-EM and biophysics core facilities for access and support, and the SuperMUC for computing time. K.-P.H. is supported by the Deutsche Forschungsgemeinschaft (CRC1064, RTG1721), the European Research Council (ERC Advanced Grant ATMMACHINE), the Gottfried-Wilhelm-Leibniz Prize and the Center for Integrated Protein Sciences Munich (CIPSM). S.E. acknowledges an EMBO long-term fellowship and K.S. acknowledges funding by Quantitative Biosciences Munich (QBM).
Reviewer information
Nature thanks B. Bartholomew, O. Llorca and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
S.E. prepared cryo-EM samples and performed structure determination. K.S. prepared nucleosomes, performed the biochemical analysis and participated in structure determination. S.E., D.K. and K.-P.H. build atomic models. K.L. characterized INO80 subcomplexes and participated in early stages of the electron microscopy analysis. M.S. operates the MPI Biochemistry cryo-EM facility, helped with electron microscopy data collection and provided general electron microscopy advice. M.M. helped with recombinant protein production. S.E. and K.-P.H. designed the overall study, analysed the results and wrote the paper with contributions from all other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
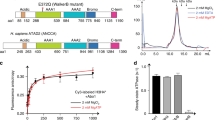
Extended Data Fig. 1 Purification of apo INO80, INO80–0N50 and sliding activity of INO80.
a, Schematic of expression and purification of INO80. b, SDS–PAGE of INO80 purification steps (stained with SimplyBlue). Protein identity was confirmed by mass spectrometry (data not shown). c, Quantification of band intensity from SDS–PAGE (SEC sample) plotted against the molecular weight shows stoichiometric presence of all subunits. d, Label-free semi-quantitative mass spectrometry analysis of INO80core complexes after individual purification steps. e, Right-angle light scattering measurement of apo INO80. Measured refractive index and calculated logarithmical molecular weight are plotted against the elution volume. The measurement yields a molecular weight of 880 kDa, confirming the integrity and correct stoichiometry of the purified complex. f, Comparison of the SEC elution profile of apo INO80 and the Arp5DBD mutant on a Superose 6 3.2/300. g, Purification of the INO80–nucleosome complex. SEC elution profile from a Superose 6 3.2/300 is shown together with an analysis of the main peak fraction by SDS–PAGE. h, Sliding of end-positioned 0N80 mononucleosomes by INO80. Native PAGE analysis of fluorescein-labelled nucleosome is shown. i, Interaction of INO80 and mononucleosome monitored by electrophoretic-mobility shift assay.
Extended Data Fig. 2 Cryo-EM data analysis.
a, b, Schemes of RELION40 classifications and refinements that were used to obtain cryo-EM reconstructions of the INO80core–NCP complex. a, Outline of an initial classification scheme that used a cryoSPARC42 ab initio 3D reconstruction of the complex as a reference. b, Classification scheme that yielded the final cryo-EM reconstructions. In a and b, boxed 3D classes were selected for further processing as indicated. Two-dimensional classes discarded for further processing are marked with an asterisk. c, Ab initio 3D reconstruction by cryoSPARC42 using 6,000 semi-automatically picked particles d, Eight hundred manually picked particles were used to obtain initial 2D classes that were used as references for automated particle picking as indicated in a. e, Projections of the experimentally determined 5.8 Å cryo-EM reconstructions obtained from the scheme in a. These projections were low-pass filtered to 35 Å and used then as templates to improve automated picking of particles corresponding to sparsely populated orientations of the complex (see Methods). The quality of the automated particle picking was verified by visual inspection of micrographs as well as by diagnostic 2D classifications (not shown). Later 3D classifications in the scheme shown in b were facilitated by masks and fixed Euler angles from previous refinements as indicated (3D classification B3). f, Gold standard Fourier shell correlation curves of final maps (3.75, 4.34, 4.62 and 4.68 Å). The resolutions were determined using the 0.143 Fourier shell correlation criterion as indicated by the dotted line. Extended Data Table 1 summarizes data collection and processing.
Extended Data Fig. 3 Cryo-EM data quality.
a, Two representative micrographs of the set that was used to determine the structure of the INO80core–NCP complex. b, Typical 2D class averages of the INO80core–NCP complex. Note that dynamic extranucleosomal DNA (extra-nuc DNA) visibly protrudes from the well-ordered core complex. c–e, The final 4.3 Å (c, overall), 3.7 Å (d, Rvb1/Rvb2–Arp5 mask) and 4.6 Å and 4.7 Å (e, grappler conformations A (right) and B (left)) maps were analysed by using ResMap50. Local resolution estimates are shown as a colour-coded surface representation along with representations of angular distributions of particles contributing to the 4.3 and 3.7 Å maps. f–m, Representative examples of cryo-EM map areas used for model building. f, The 3.7 Å map using the colour codes of Fig. 1c showing the definition of Rvb1/Rvb2–client interactions. g, ‘Explosion’ figure of the Rvb1/Rvb2 layers, along with corresponding regions of the 3.7 Å map. h, Top, details showing a representative ATP/ADP-binding site of Rvb1/Rvb2 with highlighted ADP, and showing the latch of the Ino80insert (red). i, Map area at the Arp5core showing the N-terminal brace (left), with representative details of the actin core (middle) and the ATP-binding site (right). j, Overview showing Ies6 (left) and details of its HIT-like domain (right). k, Map area at the Ies2–Rvb1/Rvb2 interaction (left) with details showing an anchoring tryptophane. l, Map area at the NCP. m, Map area at the Ino80 motor domain bound to SHL −6 (red).
Extended Data Fig. 4 Comparison of nucleosome-bound Swi2/Snf2-type ATPases.
a, Interaction of Ino80ATPase bound to SHL −6 (left, this study), Snf2ATPase bound to SHL + 6 (middle), Snf2ATPase bound to SHL + 2 and Chd1ATPase bound to SHL + 2 (right) with NCPs. b, Comparison of domain architectures of the Swi2/Snf2-type ATPases and their interaction with nucleosomal DNA.
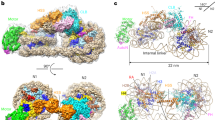
Extended Data Fig. 5 Details of Rvb1/Rvb2–Ino80insert interactions.
a, Close-up views of Rvb1 client cavities (blue), bound to the different interaction elements of Ino80insert (red, with yellow hydrophobic and green aromatic side chains). b, As in a but depicting Rvb2 client cavities. c, Ino80insert shown in rainbow colouring from N terminus (red) to C terminus (blue), to highlight the circular fold. Selected elements as well as the positions of the Rvb1/Rvb2 binding partners are annotated. d, As in c but viewed from the side to highlight the protruding plug and latch elements. e, f, Rvb1/Rvb2 pair (the pair 1c and 2c from the hexamer in Fig. 1c) bound to Ino80insert (e) compared with a Rvb1/Rvb2 pair from the unliganded dodecameric state (f) (PDB code: 4WVY). The comparison shows how client binding arranges the AAA+, OB and middle layers and displaces the N-terminal domain of Rvb1 from the client pocket, also seen for human INO80core 23. Both types of conformational changes have an effect on the ADP-binding site (ADP and ATP represented by colour-coded spheres), which suggests how client interactions are allosterically coupled to the ATPase activity of Rvb1/Rvb2. g, Exemplary view of the ADP coordination along with the superimposed map.
Extended Data Fig. 6 Two conformations of the grappler element and location of the post-HSA domain.
Masked 3D classifications identified two conformations of the grappler element of INO80core and the post-HSA domain of the Ino80ATPase. a, Left, grappler conformation A (conformation discussed in this study). Right, open conformation B in which the bar interacts with SHL –1 of the nucleosome. b, Subclass showing the post-HSA domain (magenta) at the Ino80ATPase (red). Post-HSA domain protrudes towards extranucleosomal DNA. Ies2 is depicted in orange. c, Hidden Markov model (HMM) sequence logo of Ies2, showing high sequence conservation at key Ino80 and Rvb1/Rvb2 interaction sites. d, Detailed view of the map around post-HSA domain and extranucleosomal DNA, with superimposed models.
Extended Data Fig. 7 Analysis of the enzymatic activity of INO80.
a, Sequence alignment of H2A and H2A.Z. Olive, residues at the interface of H2A with the foot of the grappler differ in a species-conserved fashion from H2A.Z. b, Sliding of 0N80 mononucleosomes by INO80 analysed by native PAGE. In the Arp5DBD mutant, K88, R90, R92, K96, R112 and R118 are mutated to alanines. AcPatch (E61A, E64A, D72A and D90A) and H2AmutZ (N73L and N89G) describe mutants of grappler-contacting residues of H2A (see Fig. 3). Individual data points with exponential fit (n = 3, technical replicates). c, Evaluation of the sliding activity of INO80. Band intensities of remodelled and unremodelled nucleosome species were quantified and the fraction of remodelled nucleosome plotted against time. Data points were fitted using an exponential equation. d, Raw data of ATPase assays. Basal ATPase rates were determined for INO80 wild type (WT) and the Arp5DBD mutant, along with nucleosome-stimulated rates. Superscripted text indicates whether a nucleosome was used to stimulate ATPase activity, and if so what type of nucleosome was used. e, ATPase rates of INO80 with and without stimulation by nucleosomes. Rates were calculated from the linear area of the raw data and were corrected for a buffer blank (colour code as in d). Mean and individual data points (n = 3, technical replicates). f, Initial sliding rates of INO80 and mutants (colour code as in c). Data were derived from exponential fits of individual sliding curves in c. Mean and individual data points (n = 3, technical replicates). g, Quotient of the sliding rate in f and ATPase rate in e normalized to the wild type. Mean and individual data points (n = 3, technical replicates).
Supplementary information
Supplementary Information
This file contains Supplementary Figure 1
Rights and permissions
About this article
Cite this article
Eustermann, S., Schall, K., Kostrewa, D. et al. Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature 556, 386–390 (2018). https://doi.org/10.1038/s41586-018-0029-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0029-y
This article is cited by
-
Energy-driven genome regulation by ATP-dependent chromatin remodellers
Nature Reviews Molecular Cell Biology (2024)
-
DNA damage and repair in the nucleosome: insights from computational methods
Biophysical Reviews (2024)
-
Identification and characterization of sugar-regulated promoters in Chaetomium thermophilum
BMC Biotechnology (2023)
-
Small molecule modulators of chromatin remodeling: from neurodevelopment to neurodegeneration
Cell & Bioscience (2023)
-
Hierarchical TAF1-dependent co-translational assembly of the basal transcription factor TFIID
Nature Structural & Molecular Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.